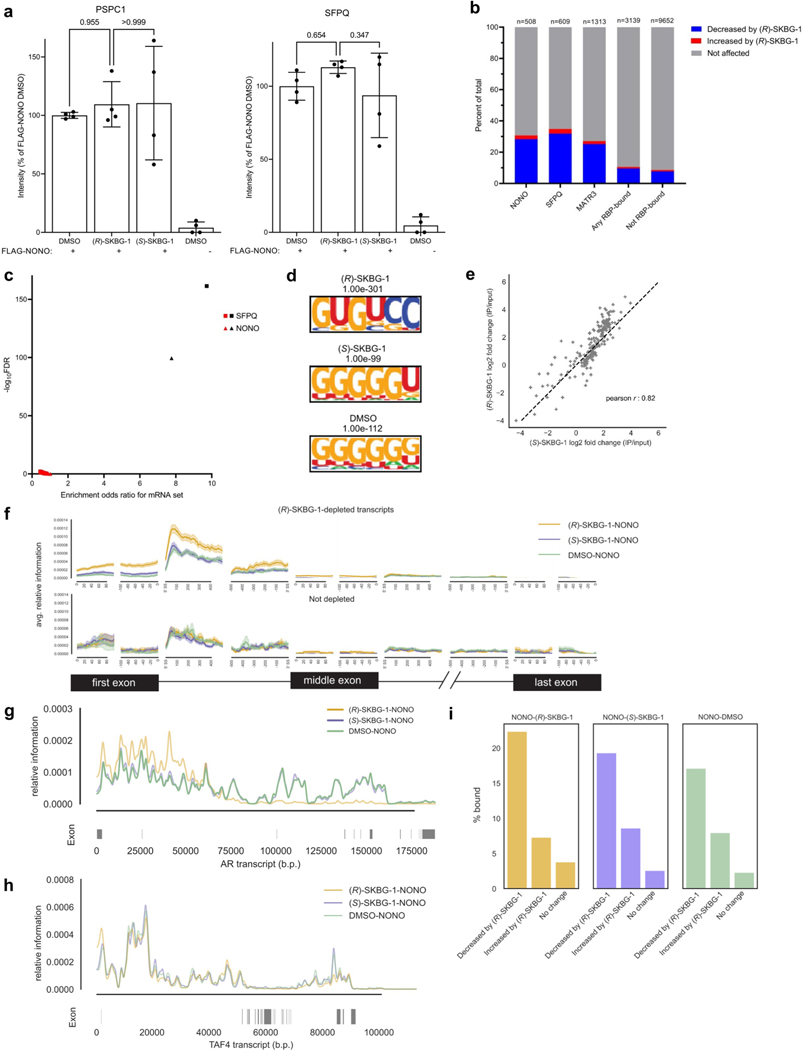
Extended Data Fig. 9 |. Effects of NONO ligands on NONO-protein and NONO-mRNA interactions.
a, Quantification of PSPC1 and SFPQ from anti-FLAG immunoprecipitation-MS experiments of FLAG-NONO-expressing cells treated with (R)-SKBG-1 or (S)-SKBG-1 (20 μM, 4 h). Data are mean values ± s.e.m.for four independent replicates. b, (R)-SKBG-1 effects on the abundance of transcripts previously shown in eCLIP experiments to bind NONO or NONO-associated RBPs (SFPQ, MATR3) versus transcripts bound to any RBP or not bound to RBPs45,46. Transcript abundances determined by RNA-seq of 22Rv1 cells treated with (R)-SKBG-1 or (S)-SKBG-1 (20 μM, 4 h). Transcripts stereoselectively decreased or increased by (R)-SKBG-1 determined as described in Fig. 3a. A bound transcript was defined as having ≥ 1 eCLIP IDR peak for a given RBP. c, Volcano plot comparing enrichment of mRNAs stereoselectively decreased by (R)-SKBG-1 in 22Rv1 cells among NONO and SFPQ-bound transcripts (in black) to 30 sets of randomly selected transcripts of matched number and expression levels (in red). Enrichment odds ratio and FDR values were determined as in Fig. 5c. d, Motifs enriched in NONO eCLIP-seq peaks from cells treated with (R)-SKBG-1 or (S)-SKBG-1 (20 μM, 4 h) or DMSO. e, Scatter plot showing repetitive element distribution between (R)-SKBG-1 and (S)-SKBG-1-treated eCLIP-seq. f, eCLIP-seq relative information content determined for transcripts bound to NONO in 22Rv1 cells treated with (R)-SKBG-1 or (S)-SKBG-1 (20 μM, 4 h) that were stereoselectively decreased or not decreased by (R)-SKBG-1. g, h, eCLIP-seq reads for total mRNA for AR (g) or TAF4 (h) in 22Rv1 cells treated with (R)-SKBG-1 or (S)-SKBG-1 (20 μM, 4 h). TAF4 is a representative mRNA that was bound to NONO but not altered in abundance by (R)-SKBG-1. For f–h, relative information content was determined as described in Fig. 5e. i, Percent of transcripts from the indicated categories bound to NONO in 22Rv1 cells treated with (R)-SKBG-1 or (S)-SKBG-1 (20 μM, 4 h) or DMSO. d-i, results are from a single eCLIP-seq experiment representative of two independent replicates.