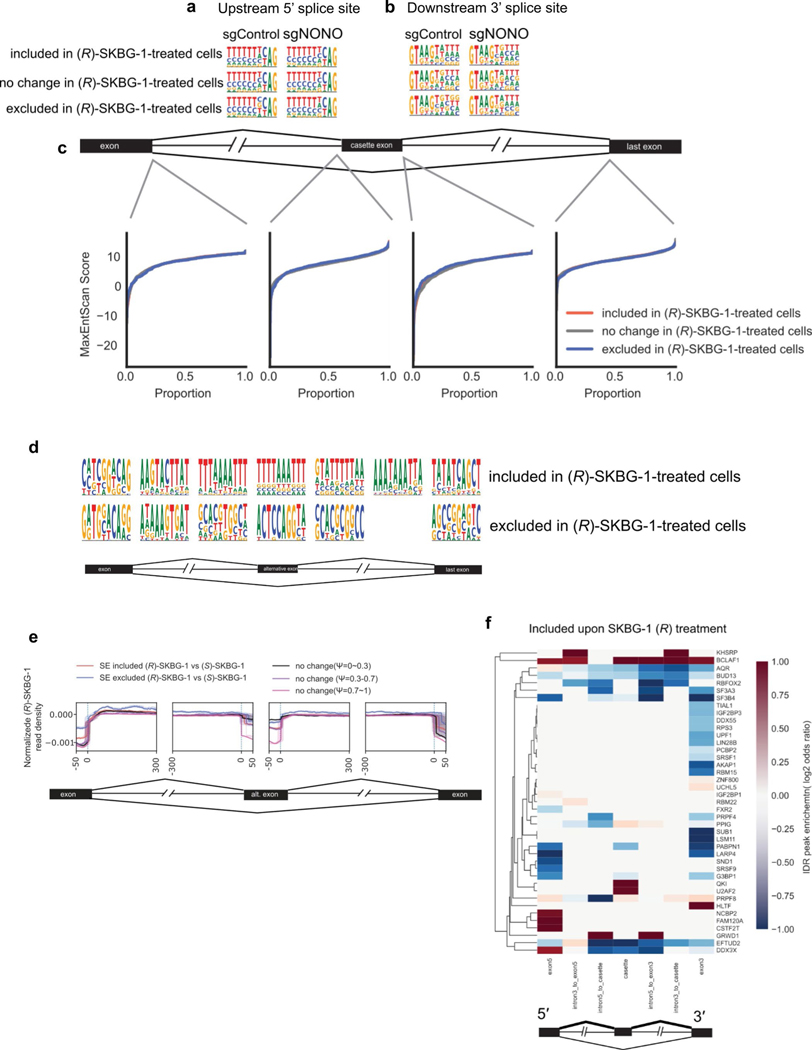
Extended Data Fig. 10 |. Effects of NONO ligands on mRNA splicing.
a, b, 5’ splice site sequence upstream (a) and 3’ splice site sequence downstream (b) of cassette exons in different contexts. Included, excluded or no change refers to the effect on cassette exons upon (R)-SKBG-1 treatment compared to (S)-SKBG-1 treatment in sgControl or sgNONO cells. c, Splice site strength of the splice sites surrounding the alternatively-spliced exons was determined using MaxEntScore. No difference was observed comparing the included/excluded group to the no change group. d, Motif enrichment comparing inclusion/exclusion events following (R)-SKBG-1 treatment to cassette exons with no change, spread across surrounding regions. Intronic sequences are defined as 500 b.p. surrounding the splice site e, Normalized eCLIP density around alternatively spliced exons for transcripts bound to NONO in 22Rv1 cells under the three indicated treatment conditions (DMSO or (R)-SKBG-1 or (S)-SKBG-1 (20 μM, 4 h treatment)), and control exons with various inclusion levels (ψ). f, Overlapping ENCODE HepG2 eCLIP IDR peaks with regions near cassette exons included after (R)-SKBG-1 treatment. Coloring shows the log2 (odds ratio) comparing included exons to regions with no change.