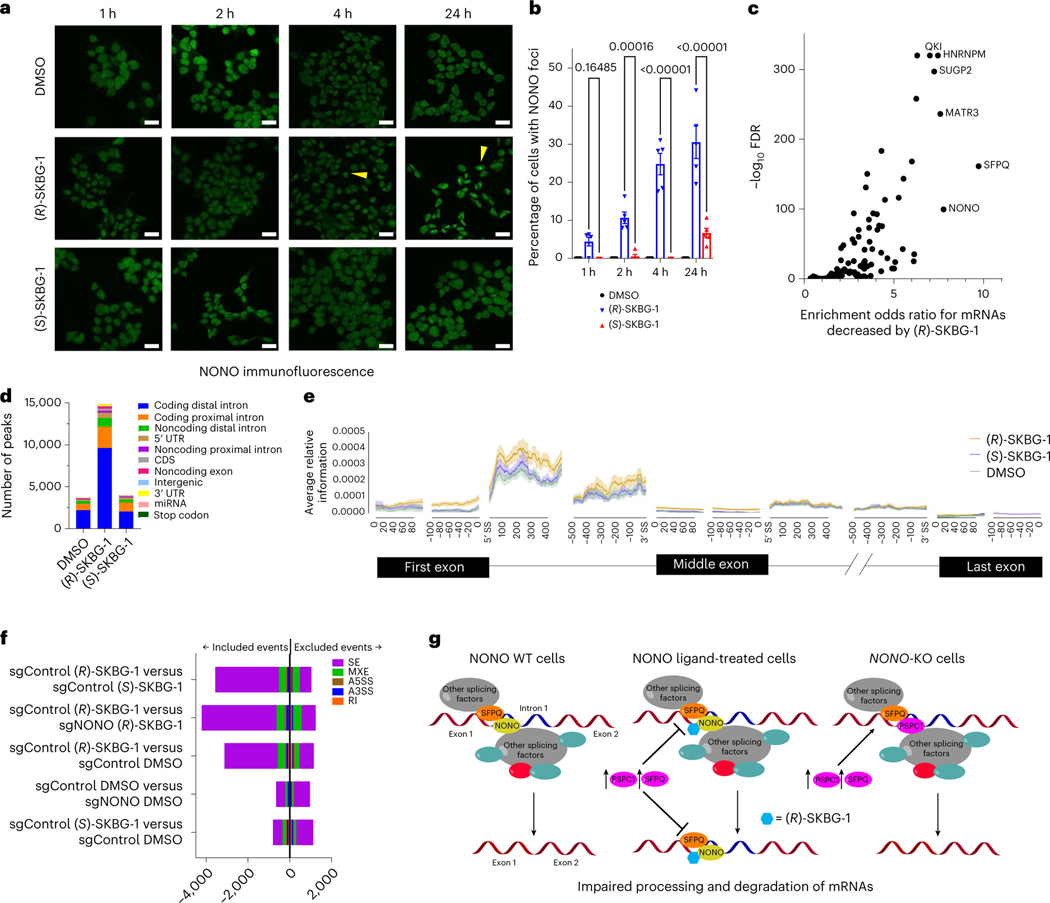
Fig. 5 |. Effects of (R)-SKBG-1 on NONO localization and RNA interactions.
a, Time-dependent effects of (R)-SKBG-1 and (S)-SKBG-1 (20 μM) on the localization of NONO in nuclear foci (yellow wedges) in 22Rv1 cells. NONO was imaged with a primary antibody (Bethyl Laboratories) and an Alexa 488 secondary antibody; scale bar, 20 μm. b, Quantification of NONO nuclear foci induced by (R)-SKBG-1 and (S)-SKBG-1. The percentage of cells with NONO foci was determined manually in a blinded manner. Data are shown as mean values ± s.e.m. for five representative images from two independent experiments where 17–124 cells (median: 58 cells) were analyzed per image. c, Volcano plot comparing enrichment of mRNAs stereoselectively decreased by (R)-SKBG-1 in 22Rv1 cells (as described in Fig. 3a) among the RNAs interacting with 104 RBPs in previous eCLIP experiments performed in HepG2 cells45,46. A bound transcript was defined as ≥1 eCLIP IDR peak for a given RBP. d,e, eCLIP–seq significant peak quantity and regional distribution (d) and relative information content (e) determined for transcripts bound to NONO in 22Rv1 cells treated with (R)-SKBG-1 and (S)-SKBG-1 (20 μM, 4 h). Relative information defined as , where and are the fractions of total reads of a given transcript in the NONO immunoprecipitation and SM-Input, respectively, that map to element . Results are from a single experiment representative of two independent experiments; UTR, untranslated region; CDS, coding sequence; miRNA, microRNA; SS, splice site. f, Effects of (R)-SKBG-1 and (S)-SKBG-1 (20 μM, 4 h) on alternative splicing events in sgControl and sgNONO 22Rv1 cells, as determined by RNA-seq. Bars indicate whether the alternative splicing event was included or excluded, respectively, by the indicated first condition compared to the second condition listed on the y axis. Significant alternative splicing events had an FDR of <0.1 and an | InclusionLevelDifference | of >0.05 (n = 3); SE, skipped exons; MXE, mutually exclusive exons; A5SS, alternative 5′ splice site; A3SS, alternative 3′ splice site; RI, retained intron. g, Trapping model for how covalent ligands targeting NONO C145 affect mRNA processing in cancer cells and subvert the compensatory action of paralogous proteins PSPC1 and SFPQ.