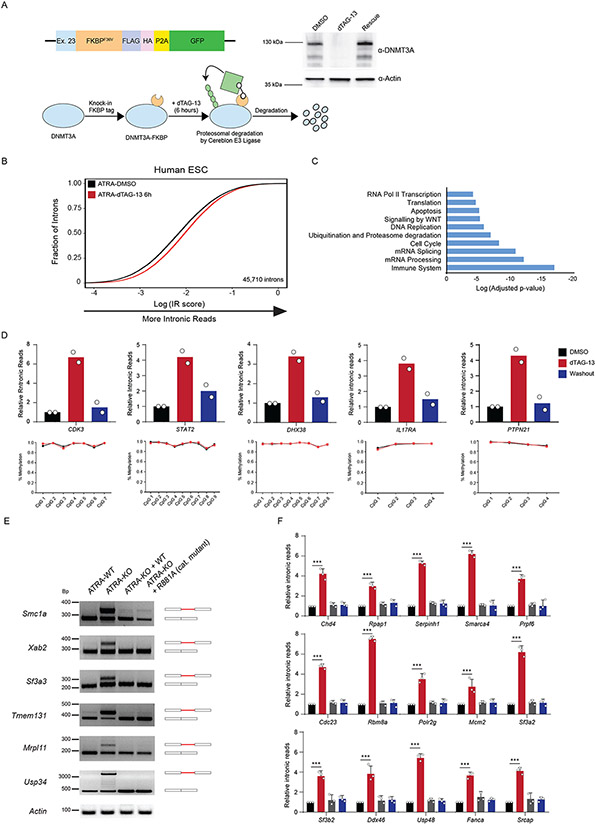
Figure 4: DNA methylation by DNMT3A is not required to rescue mis-splicing in activated stem cells.
(A) Schematic of degron construct, the degradation strategy and western blot validation after rapid degradation of FKBP-tagged DNMT3A using dTAG-13 in human ESCs (hESCs). Homology template containing an FKBP tag was introduced at the endogenous DNMT3A loci with CRISPR. Upon addition of dTAG-13, FKBP-tagged DNMT3A recruits E3 Ligase, Cereblon to polyubiquitinate and signal DNMT3A for degradation through the ubiquitin proteasome system. Western blot of DNMT3A before and after treatment with dTAG-13 for 6 hours. Washout was performed by replacing media and culturing in fresh media without dTAG-13 for an additional 12 hours. (B) eCDF for 45,710 introns in ATRA-DMSO and ATRA-dTAG-13 hESCs (n = 2 biologically independent samples) after 12 hours of treatment with dTAG-13 followed by ATRA-stimulation. Data represents an integration of all samples. (C) Gene Ontology enrichment analysis of mis-spliced genes in ATRA-dTAG-13 compared to ATRA-DMSO hESCs. (D) (top) qRT-PCR of loci displaying increased intronic reads (IR) in human ES cells (hESCs) six hours after degradation and ATRA-stimulation of FKBP-tagged DNMT3A and rescue of DNMT3A expression. (bottom) Amplicon bisulphite sequencing of intron-retained loci after rapid degradation of DNMT3A in ATRA-dTAG-13 hESCs. Bisulfite primers were created to cover all CpGs in introns. Genomic localization of each intron is: CDK3 (chr17:73998223-73998329), STAT2 (chr12:56740408-56740603), DHX38 (chr16:72144976-72146312), IL17RA (chr22:17582933-17583029), and PTPN21 (chr14:88936077-88936266). (E) Reverse Transcriptase-PCR (RT-PCR) of mis-spliced loci in ATRA-WT, ATRA-KO, and ATRA-KO after rescue with exogenous WT and catalytically inactive mutant R881A, mouse DNMT3A. (F) RT-qPCR of 15 additional mis-spliced genes (n = 3 biologically independent samples and 2 independent experiments, error bars represent mean +/− SD).