Extended Figure 9: LORALS pipeline allele specific analysis filter setting.
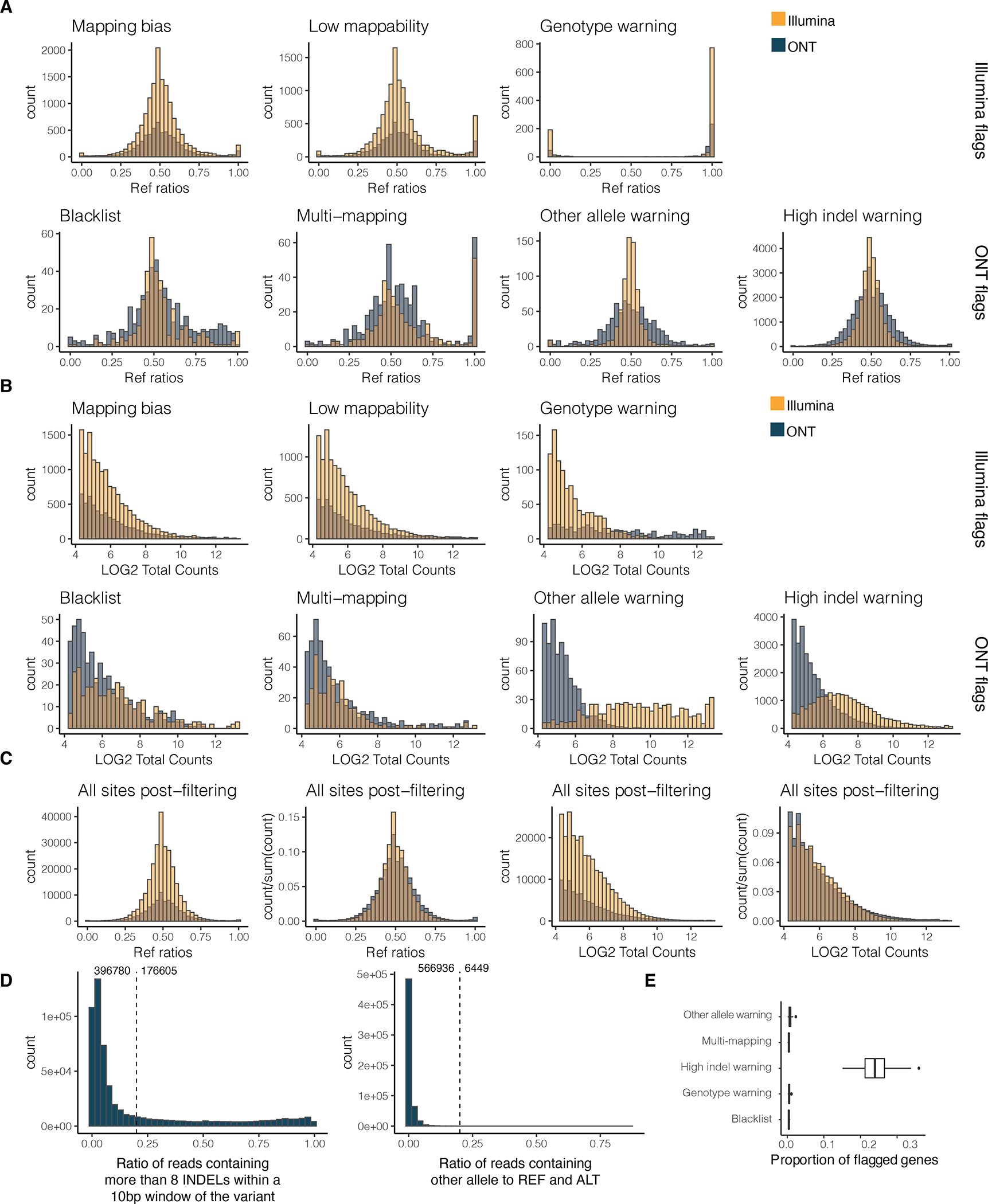
A) Reference ratio and B) normalised reads counts across different Illumina and ONT flags for both of these sequencing technologies. Mapping bias: mapping bias in simulations; Low mappability: low-mappability regions (75-mer mappability < 1 based on 75mer alignments with up to two mismatches based on the pipeline for ENCODE tracks and available on the GTEx portal); Genotype warning: no more reads supporting two alleles than would be expected from sequencing noise alone, indicating potential genotyping errors (FDR < 1%); Blacklist: ENCODE blacklist. Multi-mapping: regions with multi-mapping reads constructed using the alignability track from UCSC using a threshold of 0.1 (so that a 100kmer aligning to that site aligns to at least 5 other locations in the genome with up to 2 mismatches); Other allele warning: regions where the proportion of ref or alt containing reads is lower than 0.8; High indel warning: sited where the proportion of non-indel containing is lower than 0.8. C) Reference ratios and normalised reads counts of all kept sites across Illumina and ONT sequencing technologies. D) Distribution of the high indel warning ratios and the other allele ratios across all samples. E) Proportion of genes with at least 20 overlapping reads flagged per filter. The proportion was calculated across all genes for each sample . The center corresponds to the median, the lower and upper hinges correspond to the 25th and 75th percentiles and the whiskers extend from the hinge to the smallest/largest value no further than 1.5 * inter-quartile range from the hinge.
