Figure 2: Discovery of new transcripts and comparison between tissues.
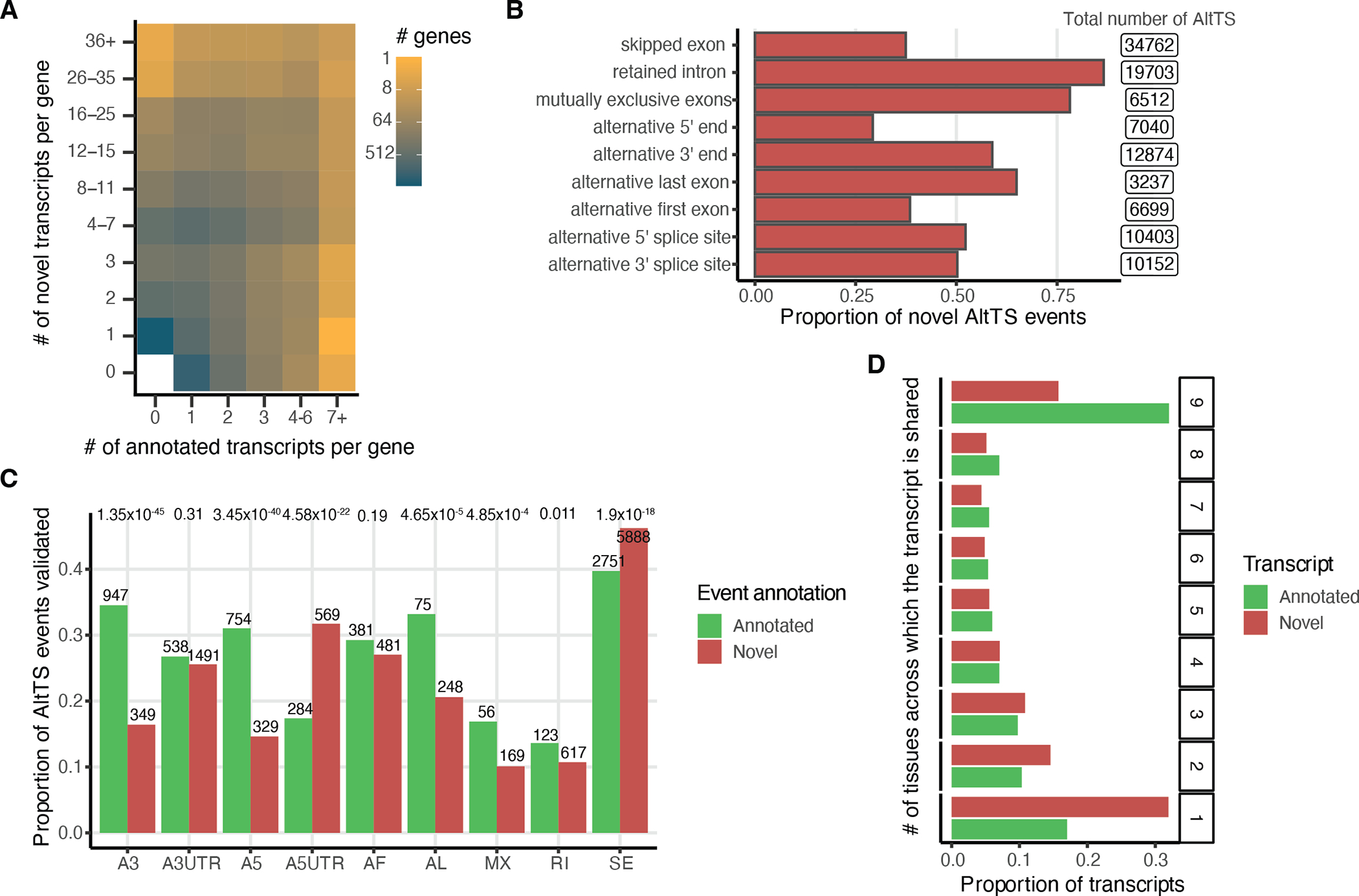
A) Number of annotated and novel transcripts per gene quantified in our dataset. B) Proportion of novel alternative transcript structure (AltTS) events across all quantified transcripts compared to GENCODEv26. C) Proportion of the AltTS events validated at the protein level by mass-spectrometry per novel or annotated. Enrichment was calculated using a two-sided proportionality test. D) Number of transcripts expressed at > 1 TPM in at least two samples and classified based on how many tissues express the transcript. A3: alternative 3’ splice site; A5: alternative 5’ splice site; AF: alternative first exon; AL: alternative last exon; A3UTR: alternative 3’ end; A5UTR: alternative 5’ end; MX: mutually exclusive exons; RI: retained intron; SE: skipped exon.
