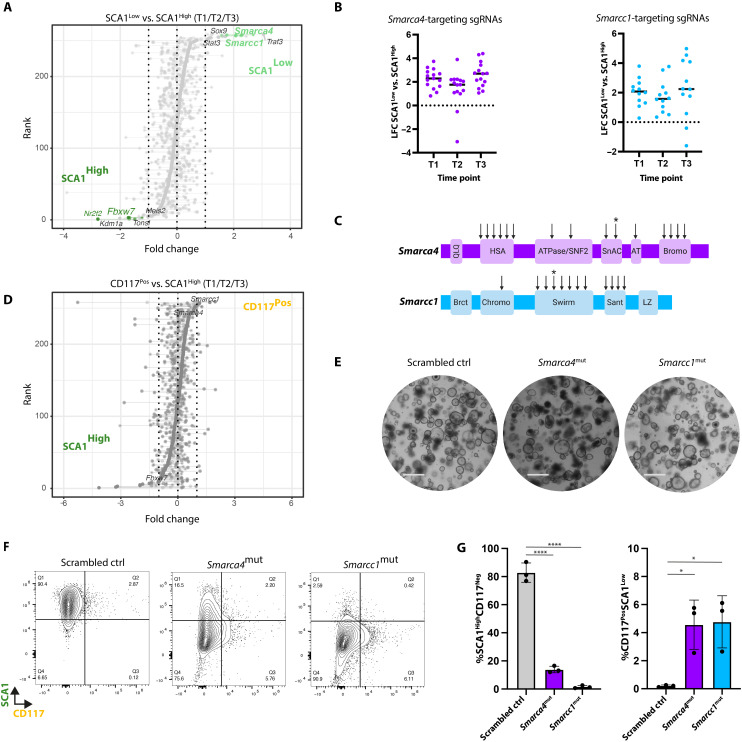
Fig. 3. Mutations in Smarca4 and Smarcc1 induce transition toward an adult-like state.
(A) Rank score plot of genes targeted by individual sgRNAs based on their FC in the SCA1LowCD117Neg versus SCA1HighCD117Neg gate for all three time points versus T = 0. x axis: FC; y axis: rank score according to the combined effect of all sgRNAs targeting the indicated genes. Gene names indicated the top 5 ranked genes in the SCA1High (dark green) and SCA1Low (light green) populations. (B) Dot plot of the LFC of sgRNAs found in SCA1LowCD117Neg versus SCA1HighCD117Neg gates targeting Smarca4 (left) and Smarcc1 (right) across the three time points. x axis: time point; y axis: LFC of the normalized sgRNA abundance in SCA1LowCD117Neg versus SCA1HighCD117Neg gates. (C) Schematics of the functional domains targeted by sgRNAs (arrows) for Smarca4 (top) and Smarcc1 (bottom). Asterisks indicate sgRNAs used for the generation of mutant lines for validation experiments. (D) Rank score plot of genes targeted by individual sgRNAs based on their FC in the CD117Pos versus SCA1HighCD117Neg gate for all three time points versus T = 0. x axis: FC, y axis: rank score according to the combined effect of all sgRNAs targeting the indicated genes. Genes of interest are indicated by name. (E) Bright-field images of FEnS representing the scrambled control line and lines carrying mutations in Smarca4 and Smarcc1. Scale bars, 1 mm. (F) Flow cytometry contour plots showing CD117 and SCA1 expression in scrambled control, Smarca4mut, and Smarcc1mut fetal organoids. (G) Bar plot of percentage of SCA1HighCD117Neg (left) and SCA1LowCD117Pos (right) cells in scrambled control, Smarca4mut, and Smarcc1mut FEnS. n = 3, error bars indicate SD, unpaired Student’s t test. *P < 0.05 and ****P < 0.0001.