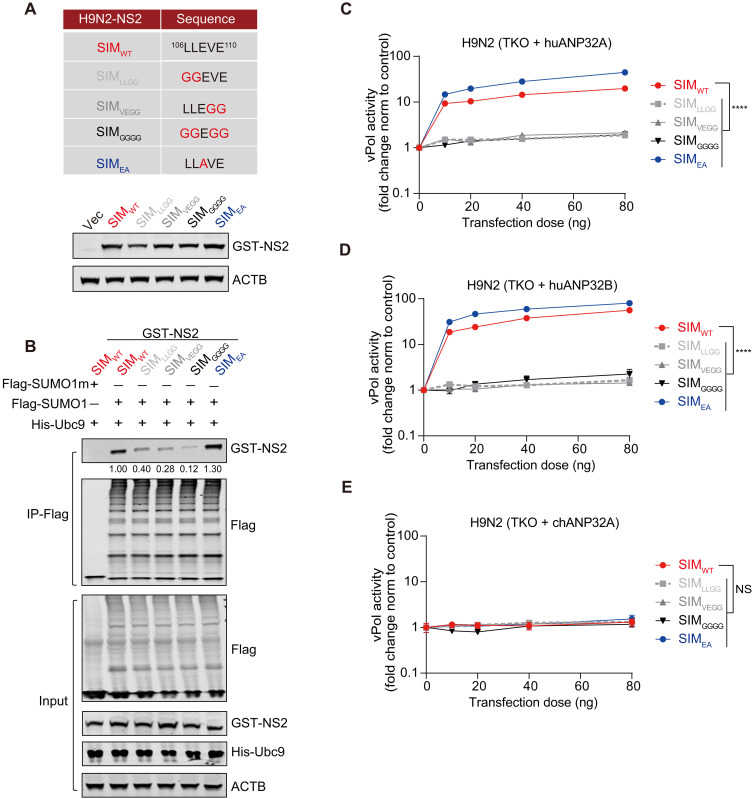
Fig. 7. The SIM in NS2 determines its avian-signature polymerase-enhancing function.
(A) Panel of generated NS2 mutants. The accompanying WBs show the effect of NS2 SIM changes on protein expression levels in these mutants. HEK293T cells were transfected with equal amount of indicted GST-tagged H9N2-NS2 constructs. At 24 hours after transfection, cell lysates were prepared with radioimmunoprecipitation assay lysis buffer and subjected to SDS–polyacrylamide gel electrophoresis, followed by analysis with Western blotting. (B) Effects of H9N2-NS2 SIM mutations on H9N2-NS2 binding to Flag-tagged SUMO1. Experiments were performed as in Fig. 6E. Three independent experiments were performed, with comparable results in each experiment. (C to E) vPol reconstitution assay to compare the effect of H9N2-NS2 SIM mutations on H9N2 vPol activity in TKO cells reconstituted with huANP32A (C), huANP32B (D), and chANP32A (E). TKO cells were transfected with H9N2-PB2, H9N2-PB1, H9N2-PA, H9N2-NP, different ANP32s as indicated, a mini-genome reporter, and a Renilla expression control together with increasing dose of different H9N2-NS2. Luciferase activity was measured 24 hours later. For all assays, data were firefly activity–normalized to Renilla and plotted as fold change to empty vector (0 ng of NS2 constructs). Data are presented as the means ± SD (n = 3) and are one representative of three independent experiments. Significance was determined by two-way ANOVA. ****P < 0.0001.