Abstract
Meta-diamides (e.g. broflanilide) and isoxazolines (e.g. fluralaner) are novel insecticides that target the resistant to dieldrin (RDL) subunit of insect γ-aminobutyric acid receptors (GABARs). In this study, we used in silico analysis to identify residues that are critical for the interaction between RDL and these insecticides. Substitution of glycine at the third position (G3’) in the third transmembrane domain (TMD3) with methionine (G3’M TMD3), which is present in vertebrate GABARs, had the strongest effect on fluralaner binding. This was confirmed by expression of RDL from the rice stem borer, Chilo suppressalis (CsRDL) in oocytes of the African clawed frog, Xenopus laevis, where the G3’MTMD3 mutation almost abolished the antagonistic action of fluralaner. Subsequently, G3’MTMD3 was introduced into the Rdl gene of the fruit fly, Drosophila melanogaster, using the CRISPR/Cas9 system. Larvae of heterozygous lines bearing G3’MTMD3 did not show significant resistance to avermectin, fipronil, broflanilide, and fluralaner. However, larvae homozygous for G3’MTMD3 were highly resistant to broflanilide and fluralaner whilst still being sensitive to fipronil and avermectin. Also, homozygous lines showed severely impaired locomotivity and did not survive to the pupal stage, indicating a significant fitness cost associated with G3’MTMD3. Moreover, the M3’GTMD3 mutation in the mouse Mus musculus α1β2 GABAR increased sensitivity to fluralaner. Taken together, these results provide convincing in vitro and in vivo evidence for both broflanilide and fluralaner acting on the same amino acid site, as well as insights into potential mechanisms leading to target-site resistance to these insecticides. In addition, our findings could guide further modification of isoxazolines to achieve higher selectivity for the control of insect pests with minimal effects on mammals.
Author summary
Meta-diamides (e.g. broflanilide) and isoxazolines (e.g. fluralaner) are members of group 30 of compounds according to the Insecticide Resistance Action Committee, and are defined as γ-aminobutyric acid (GABA)-gated chloride channel allosteric modulators. However, their mode of action on the GABAR remains to be fully elucidated. In the current study, we provide in silico, in vitro and in vivo evidence that G3’TMD3, the glycine residue at the third position (G3’) in the third transmembrane domain (TMD3) of the insect GABAR subunit, resistant to dieldrin (RDL), is critical for the action of these insecticides. Furthermore, the homozygous but not heterozygous lines of the fruit fly Drosophila melanogaster bearing G3’MTMD3 showed high resistance to broflanilide and fluralaner but were unable to survive to the pupal stage. These findings enhance our understanding of resistance-related and homozygous-lethal gene mutations, and may prove useful for the future synthesis of related insecticides to gain long-term effectiveness and higher selectivity.
Introduction
High and efficient agricultural activity is required to meet the demand of an ever-growing human population. However, agricultural productivity is hampered by insect pests, which can lead to 20%-30% loss of crops [1]. To date, insecticides are still the most widely used tool for controlling insect pests. However, crop protection efforts are undermined by the development of resistance to insecticides. Exploring the molecular targets of insecticides forms an important basis for understanding mechanisms leading to resistance [2].
The insect γ-aminobutyric acid receptor (GABAR) subunit, RDL (resistant to dieldrin), is the molecular target for various types of insecticides, such as cyclodienes, phenylpyrazoles, macrocyclic lactones, meta-diamides and isoxazolines [3] (S1 Fig). Meta-diamides and isoxazolines are novel classes of compounds, which have been classified into group 30 by the Insecticide Resistance Action Committee, and are used to protect crops and animals from insect pests [4]. Despite acting on the same molecular target, meta-diamides and isoxazolines do not show cross-resistance with cyclodienes or phenylpyrazoles [3]. Previous studies demonstrated that mutations at A302 (the fruit fly, Drosophila melanogaster, numbering, otherwise referred to as A2’TMD2) in the second transmembrane domain (TMD2) of RDL underlie resistance to fipronil and dieldrin [3,5,6]. In contrast, the potency of meta-diamides and isoxazolines is unaffected by mutations at A2’TMD2, suggesting that they act on a site different to that of cyclodienes or phenylpyrazoles [7–10]. In line with this, G3’MTMD3, a mutation in TMD3 of heterologously expressed RDL, substantially reduced the potency of meta-diamides and isoxazolines (S2 Fig) [10–13]. Attempts to generate diamondback moth Plutella xylostella that were resistant to broflanilide were unsuccessful after ten generations of selection [14]. Thus, potential mechanisms underlying resistance to meta-diamides, including mutations in RDL, remain to be identified in vivo.
In the present study, differences between arthropod RDL and mammalian GABAR subunits (e.g. α, β and γ) were explored to highlight and reinforce residues in the TMDs as being important for the interaction between GABARs and meta-diamides (e.g. broflanilide) or isoxazolines (e.g. fluralaner). Also, D. melanogaster, the classical insect model organism, was edited using the CRISPR/Cas9 system to determine whether mutations at G3’TMD3 in RDL in vivo lead to resistance to broflanilide and fluralaner. Knowledge gained from this study can guide further modification of novel insecticides to achieve highly selective toxicity to insect pests with minimal effect on mammals, as well as provide insights into a potential mechanism underlying resistance to meta-diamides and isoxazolines in the field.
Results
Prediction of potential binding sites of fluralaner
Amino acid residues of arthropod RDL and vertebrate GABAR subunits were aligned and the four TMDs were identified (S3 and S4 Figs and S1 Table and S1 Note). The effect of mutant residues in the TMDs on binding of fluralaner to a three-dimensional homology model of RDL from the rice stem borer, Chilo suppressalis, was assessed. Altering several residues in RDL to the equivalent amino acid present in vertebrate GABARs increased the binding energy of fluralaner (S1 Table). Twelve of these mutations were selected for functional expression in oocytes of the African clawed frog, Xenopus laevis, in order to determine if they play a role in the interaction between RDL and fluralaner (S1 Table).
Electrophysiological responses of wild-type and mutant CsRDL to GABA and fluralaner
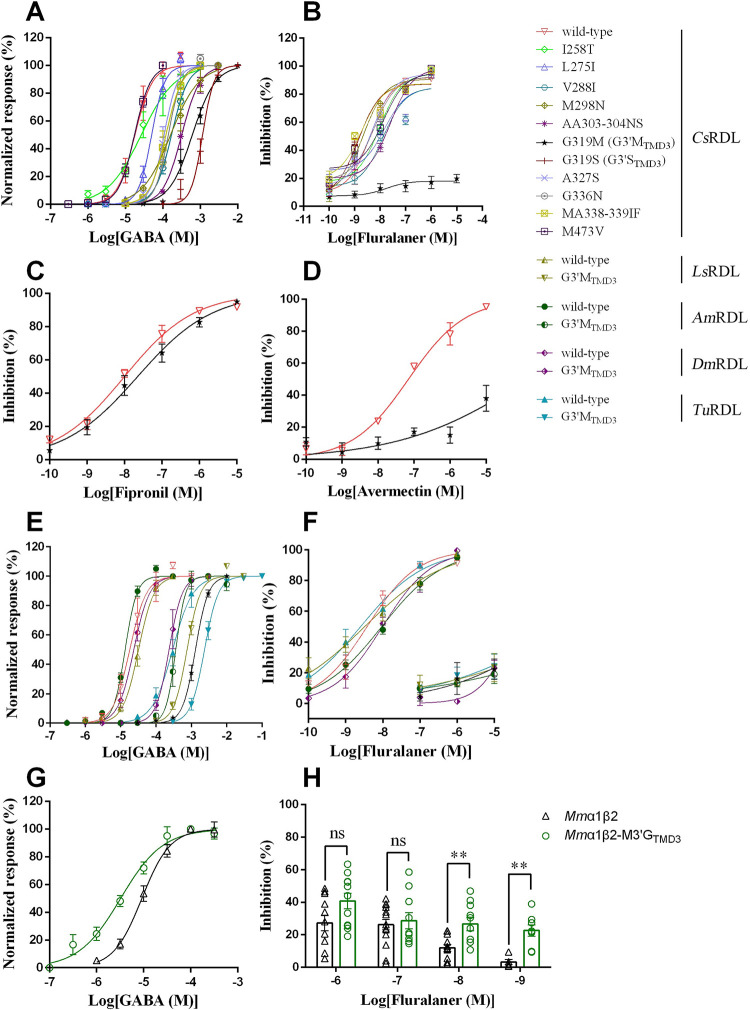
Twelve homomeric mutant C. suppressalis (CsRDL) subunits were expressed in X. laevis oocytes (S2 Table). Inward currents upon GABA application were not detected for one mutant, I477D. The maximum GABA-induced current (Imax) was notably lower in the mutant channels than that of the wild-type CsRDL channel except for G3’MTMD3. All mutations except for I258T and M473V decreased the potency of GABA as indicated by a significant increase in EC50 (Figs 1A and S5 and S2 Table). The EC50 values of G3’MTMD3 and G3’STMD3 increased 34- and 64-fold, respectively, compared with that of the wild-type CsRDL channel (Fig 1A and S2 Table).
Fig 1. Responses to GABA and antagonists in Xenopus laevis oocytes expressing wild-type and mutant arthropod or mammalian GABAR subunits.
(A) and (B) Concentration-response curves of GABA (A) and inhibition of GABA-induced currents by fluralaner (B) from wild-type or mutant RDL receptors. (C) and (D) Inhibition of GABA-induced currents by fipronil (C) and avermectin (D) in wild-type or G3’MTMD3 RDL. (E) and (F) Effect of the G3’MTMD3 mutation in RDL of different arthropod species on response to GABA (E) or fluralaner (F). (G) and (H) Concentration-response curves of GABA (G) and inhibition of GABA-induced currents by fluralaner (H) from heteromeric Mmα1β2 or mutant Mmα1β2-M3’GTMD3 channels. Error bars indicated the standard error of the mean (SE). Significant difference was determined by Student’s t-test (ns, not significant; **, P < 0.01).
Fluralaner showed concentration-dependent antagonistic action on the GABA (EC50)-induced currents (Figs 1B and S5) with an IC50 of 4.20 nM in the wild-type CsRDL channel (S3 Table). The double-mutation (AA303-304NS) caused a significant 5.91-fold increase in IC50, whilst G3’MTMD3 decreased the potency of fluralaner even further with an IC50 > 10,000 nM.
G3’MTMD3 reduces the sensitivity of RDL to avermectin
Both fipronil and avermectin strongly inhibited the GABA-induced current in the homomeric wild-type CsRDL channel with an IC50 of 10.02 and 69.90 nM, respectively (Fig 1 and S4 Table). However, the G3’MTMD3 CsRDL channel was less sensitive than the wild-type to fipronil or avermectin as indicated by significantly greater IC50 values. In particular, avermectin at 10−5 M only inhibited 38.06% of the GABA-induced response in CsRDL with the G3’MTMD3 as opposed to almost abolishing the GABA response in wild-type channels (Fig 1D).
G3’MTMD3 reduces the sensitivity of arthropod RDL to fluralaner
To verify whether G3’MTMD3 can potentially give rise to resistance to fluralaner in different arthropod species, the mutation was introduced into RDL from Hymenoptera (Apis mellifera), Hemiptera (Laodelphax striatellus), Arachnoidea (Tetranychus urticae), Lepidoptera (C. suppressalis), and Diptera (D. melanogaster). The mutant RDL subunits were individually expressed in X. laevis oocytes. In each case, G3’MTMD3 reduced the potency of GABA by 8–34-fold and showed significantly different EC50 values compared with that of the corresponding wild-type RDL (Fig 1E and S5 Table). In addition, the G3’MTMD3 in RDL of each species reduced the potency of fluralaner as shown by significantly increased IC50 values (Fig 1F and S5 Table).
M3’GTMD3 in the Mmα1β2 GABAR affects the potency of fluralaner
To examine the contribution of G3’TMD3 of mammalian GABARs towards fluralaner insensitivity, the M3’GTMD3 was introduced into the mouse Mus musculus GABAR β2 subunit (Mmβ2). There was no detectable response to GABA in X. laevis oocytes injected with Mmα1 or Mmβ2 alone. However, co-expression of Mmα1 and Mmβ2 subunits generated a functional heteromeric GABA-gated channel (Mmα1β2) (Fig 1 and S6 Table). GABA potency on the Mmα1β2-M3’GTMD3 channel was enhanced as shown by a significantly lower EC50 value compared to that of the wild-type (Fig 1G and S6 Table). Fluralaner showed decreased potency on Mmβ2-M3’GTMD3 at concentrations of 10−8 and 10−9 M as shown by significantly less inhibition of GABA-induced currents (Fig 1H and S7 Table).
Generation of genome-modified D. melanogaster bearing mutations at G3’TMD3
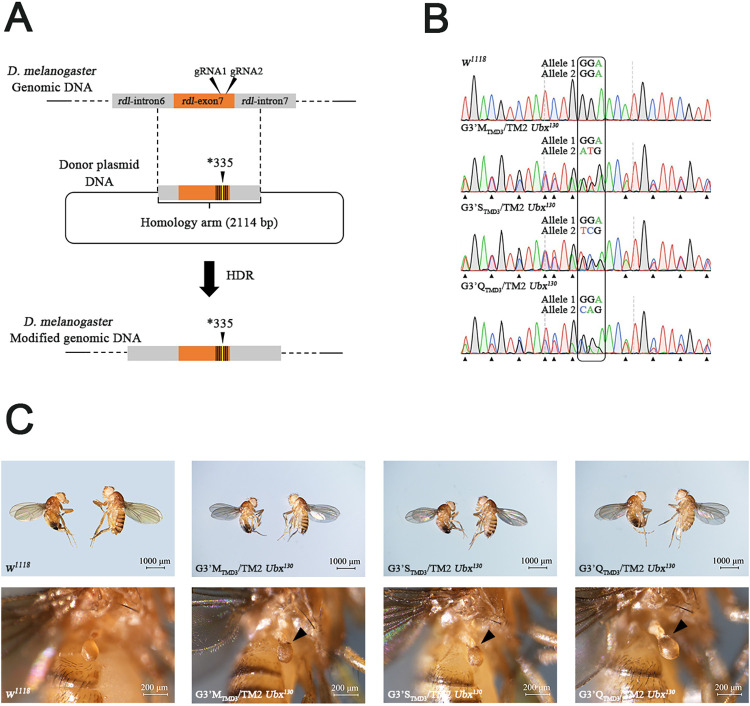
The CRISPR/Cas9 genome editing system was used to introduce substitutions at G3’TMD3 (G335) in DmRDL (Figs 2A and S6). For the G3’MTMD3 or G3’STMD3, a mixture including Cas9 mRNA, donor plasmid and gRNAs was injected into the embryos of the w1118 strain (defined as G0 generation). In the G0 generation, 2 out of 12 or 4 out of 25 individuals carrying the G3’MTMD3 or G3’STMD3 allele, respectively, were identified. Subsequently, the G1 individuals from these positive lines were crossed with the balancer strain (w1118; TM2 Ubx130/TM6B Tb1) to retain the mutant allele in the G2 generation (S7 Fig). After self-crossing of G2 generation, G3’MTMD3 or G3’STMD3 homozygous lines were not observed in the progeny of the G3 generation, indicating that either mutation at G3’TMD3 may cause homozygous-lethality in D. melanogaster adults. Therefore, strains carrying the TM2 Ubx130 balancer chromosome, which are also heterozygous for G3’MTMD3 or G3’STMD3, were generated and verified by genomic DNA sequencing (Fig 2).
Fig 2. CRISPR/Cas9-mediated point-mutation at G3’TMD3 in the Rdl gene of Drosophila melanogaster.
(A) Diagram of the genome editing strategy. Black triangles indicated the gRNA-targeted sites. A 2114 bp homologous region with a modified codon corresponding to the G3’TMD3 residue was cloned into the donor plasmid, and nine synonymous mutations indicated by vertical lines were designed around the G3’TMD3 residue to prevent repeated editing. (B) Genotypes of the heterozygous mutant strains were confirmed by sequencing of genomic DNA. The corresponding codon of the G3’TMD3 residue is boxed, and the synonymous mutations are indicated by black equilateral triangles. (C) Balancer-associated phenotype of heterozygous mutant strains. Additional bristles on the haltere are indicated by a black triangle.
Because G3’QTMD3 homozygous lines could not be generated with the w1118 strain, the nos.Cas9 strain with a different genetic background in the X chromosome was used for establishing the G3’QTMD3 homozygous lines injected with specific gRNAs/donor plasmid mixture (S6 Fig), which was defined as G0nos. The G0nos adults were individually crossed with the nos.Cas9 flies, and the G1nos progeny were identified by genomic DNA sequencing. The G3’QTMD3 mutation was detected in 1 out of 18 G1nos lines, indicating that the homology-directed repaired (HDR) allele (G3’QTMD3) was present in the G0nos strain. Unfortunately, similar to G3’MTMD3 and G3’STMD3, G3’QTMD3 was homozygous lethal. Therefore, a heterozygous G3’QTMD3 strain was used for further study (Fig 2).
In the complementation experiment, all progeny produced by these crosses carried the parental balancer chromosomes, thus no complementation was apparently viable. This result confirmed that the observed lethality was linked to the corresponding genomic region containing Rdl, presumably to G3’M/S/QTMD3. Therefore, we concluded that the substitutions at G3’TMD3 of the RDL subunit might have a severe impact on channel function affecting viability.
Heterozygous adults bearing a mutation at G3’TMD3 were not resistant to insecticides targeting the GABAR
Heterozygous D. melanogaster adults bearing a mutation at G3’TMD3 (G3’MTMD3, G3’QTMD3 or G3’STMD3) were sensitive to fipronil, fluralaner, broflanilide and avermectin without significant alteration in resistance ratio (S8 Table). The findings in the current and previous studies indicated that mutations at G3’TMD3 reduced potency of these insecticides when using in vitro and in silico approaches [10,12,15].
Temporal characteristics of lethality caused by homozygous mutations at G3’TMD3
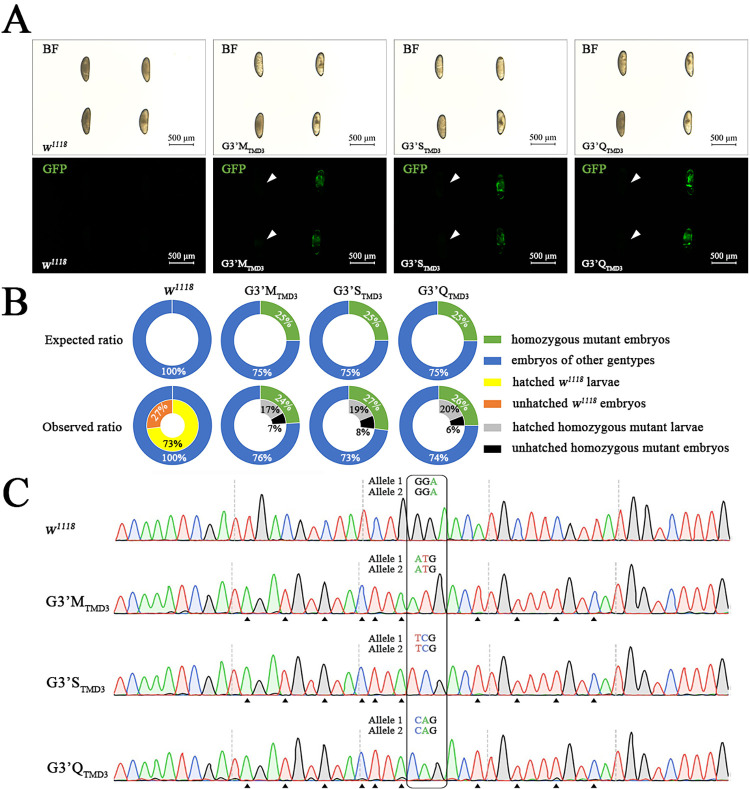
In order to further investigate the substitution-associated lethality of homozygous mutations at G3’TMD3 in RDL, we crossed the heterozygous mutants (G3’MTMD3/TM2 Ubx130, G3’STMD3/TM2 Ubx130 or G3’QTMD3/TM2 Ubx130) with the w1118; dIRE1Δ/TM3 Sb GFP strain to obtain mutants carrying a GFP-labeled balancer. Analysis of embryos showed that the GFP and homozygous mutations at G3’TMD3 in RDL did not hinder embryogenesis of D. melanogaster (Fig 3A). The proportion of embryos homozygous for G3’MTMD3, G3’STMD3 or G3’QTMD3 was 24%, 27% and 26%, respectively (Fig 3B). Furthermore, their corresponding hatching rate was 70.83%, 70.37% and 76.92%, respectively, and that of w1118 was 73.00%. These results demonstrated that the homozygous embryos of G3’MTMD3, G3’STMD3 or G3’QTMD3 could develop and hatch normally.
Fig 3. Identification of D. melanogaster embryos bearing homozygous mutations at RDL G3’TMD3.
(A) Fluorescence detection of embryos. Heterozygous mutant or TM3 Sb GFP/ TM3 Sb GFP embryos showing a GFP signal. Homozygous mutant embryos labeled with white triangles showed no GFP signal. BF, bright field. (B) The proportion and hatching rate of embryos bearing homozygous mutations. For each strain, genotypes of 100 embryos were identified. 70.83% (17 of 24), 70.37% (19 of 27) and 76.92% (20 of 26) embryos hatched that were homozygous for G3’MTMD3, G3’STMD3 and G3’QTMD3, respectively. (C) Genotypes of the homozygous mutant larvae were confirmed by sequencing of genomic DNA. The codon of the G3’TMD3 residue is boxed and the synonymous mutations are labeled with black equilateral triangles.
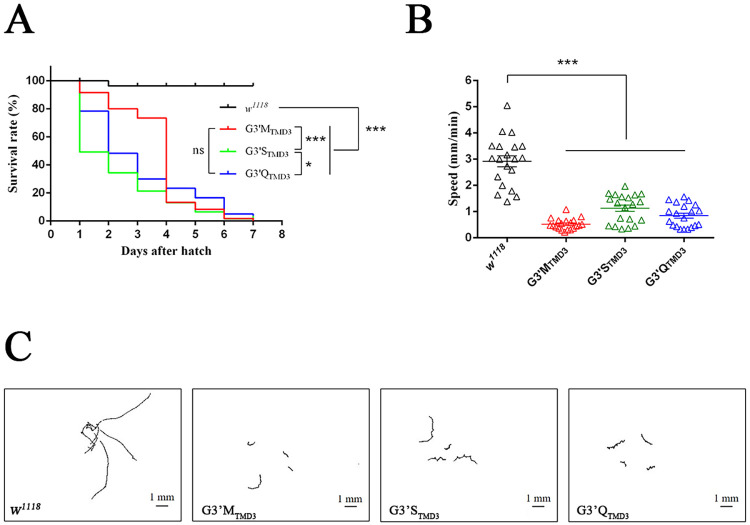
After hatching of the three homozygous G3’MTMD3 strains of D. melanogaster, a significant difference was observed in their survival rate (Fig 4A and S9 Table). Approximate 90% of their larvae survived for one day and > 70% for three days. However, only 30% and 20% of homozygous G3’QTMD3 and G3’STMD3 larvae, respectively, survived for three days. All homozygous mutant flies stayed at the larval stage until death within 7 days, whilst the control strain, w1118, progressed to pupation.
Fig 4. Lethality and reduced locomotion in D. melanogaster larvae caused by homozygous mutations at G3’TMD3.
(A) Temporal characteristics of lethality in homozygous mutants. Data were analyzed using the Log-rank test for trend and the Mantel-Cox test (ns, not significant; *, P < 0.05; ***, P < 0.001). (B) Crawling speed of homozygous mutants. Error bars indicate the standard error of the mean (SE). Significant difference was determined by Student’s t-test (***, P < 0.001). (C) Representative motion path of homozygous mutants in 2 minutes (S1 Video).
Larvae bearing the homozygous G3’MTMD3 were resistant to fluralaner and broflanilide
Because the survival rate of the homozygous G3’MTMD3 larvae was higher than 90% at 24 h after hatching (Fig 4A), the sensitivity of newly-hatched larvae towards fluralaner and broflanilide were measured. As shown in Table 1, LC50 values of fluralaner and broflanilide for the w1118 larvae were 1.08 and 0.47 mg/kg, respectively. In contrast, the mortality of the homozygous G3’MTMD3 larvae was 35.71% and 39.54% to fluralaner and broflanilide at 1000 mg/kg, respectively, which indicated that they have developed > 900-fold resistance to these two insecticides compared to the w1118 larvae. Such high levels of cross-resistance suggested that the G3’TMD3 residue plays a critical function in the interaction between RDL and fluralaner or broflanilide in vivo. In addition, it is worth noting that the homozygous G3’MTMD3 larvae also showed resistance (6.80-fold) to avermectin but not to fipronil.
Table 1. Sensitivity of the wild-type w1118 larvae and homozygous G3’MTMD3 larvae to four different insecticides.
| Insecticide | Genotype | Slope ± SE | LC50 (95% CI †) (mg/kg) | χ2 (df) | P value | RR‡ |
|---|---|---|---|---|---|---|
| Fipronil | w 1118 | 2.03±0.27 | 0.31 (0.22–0.40) | 2.51 (13) | 0.999 | 1.00 |
| G3’MTMD3 | 1.49±0.20 | 0.31 (0.20–0.44) | 4.77 (13) | 0.980 | 1.00 | |
| Avermectin | w 1118 | 2.67±0.38 | 0.10 (0.08–0.11) | 8.59 (13) | 0.803 | 1.00 |
| G3’MTMD3 | 1.63±0.24 | 0.68*(0.49–0.89) | 3.98 (13) | 0.991 | 6.80 | |
| Fluralaner | w 1118 | 1.14±0.14 | 1.08 (0.73–1.57) | 5.02 (13) | 0.975 | 1.00 |
| G3’MTMD3 | — | >1000* | — | — | > 925.93 | |
| Broflanilide | w 1118 | 0.87±0.13 | 0.47 (0.25–0.76) | 4.66 (13) | 0.982 | 1.00 |
| G3’MTMD3 | — | > 1000* | — | — | > 2127.66 |
† 95% confidence interval.
* indicates significant difference relative to the wild-type w1118 as determined by the 95% CI without overlapping.
‡ RR indicates resistance ratio: resistance ratio = LC50 value of the G3’MTMD3/LC50 value of the wild-type w1118.
Larvae homozygous for G3’MTMD3 displayed less locomotion
A crawling assay was used to investigate the potential fitness cost caused by homozygous mutations at the RDL G3’TMD3 residue. As shown in Fig 4 and S1 Video, D. melanogaster larvae homozygous for G3’MTMD3, G3’QTMD3 or G3’STMD3 showed an average speed of 0.52, 0.85 and 1.13 mm/min, respectively (S10 Table). In contrast, the wild-type w1118 larvae were faster at 2.92 mm/min (Fig 4B and 4C), indicating a significant mutation-associated decrease in locomotion.
Discussion
To date, insects have developed varying levels of resistance to traditional GABAR-targeting insecticides such as cyclodienes, phenylpyrazoles and macrocyclic lactones [5,16–18]. Previous studies investigating mechanisms of resistance have provided insights into their binding site(s). For example, A2’TMD2 of RDL is recognized as the common binding site of cyclodienes and phenylpyrazoles. A mutation at A2’TMD2 is the critical basis for resistance to dieldrin (A2’STMD2) or fipronil (A2’NTMD2) [19–23]. However, RDL with resistance-related mutations of A2’S/G/NTMD2 is still inhibited by fluralaner [24–28].
Meta-diamides (e.g. broflanilide) and isoxazolines (e.g. fluralaner) are defined as non-competitive antagonist (NCA)-II compounds that share a coupled action site in proximity to the interface of TMD1/TMD3 in RDL [3,29]. In the current study, the amino acid sequences of arthropod and mammalian GABARs were aligned in order to explore the potential binding site of fluralaner. A similar research approach was performed on the bumblebee Bombus impatiens BiNav1 sodium channel, which identified additional amino acid residues that underlie the sensitivity of B. impatiens to pyrethroids as well as selective resistance to tau-fluvalinate [30]. With mammals being relatively insensitive to fluralaner [24,31], we identified 35 amino acid residues in the TMDs that differ between arthropod and mammalian GABAR subunits (S1 Table). Heterologous expression studies showed that the G3’MTMD3 mutation led to the greatest decrease in fluralaner potency (S3 Table) reinforcing this residue as being crucial for isoxazoline activity, which is consistent with previous studies [10,12]. Meanwhile, G3’MTMD3 also considerably decreased the antagonist actions of avermectin (S4 Table). Avermectin is a macrocyclic lactone and is thought to act primarily on the glutamate-gated chloride channel (GluCl) with GABARs being a secondary target [32]. In line with this, both G323D in TuGluCl and G315E in PxGluCl (equivalent position to RDL G3’TMD3) are associated with resistance to avermectin [33,34]. Although the G3’M mutation might allosterically affect GABA binding to the orthosteric site leading to the significantly different EC50 values compared to that of wild-type (S2 Table), the concentration-response curve indicated that the channel was still able to function in response to GABA (Fig 1A and 1B). Other studies found that fluralaner [11,25] and avermectin inhibited [3H]fluralaner binding on M. domestica membranes [28]. Fluralaner binding was also inhibited when the G3’MTMD3 was present in the homomeric RDL channels of the common cutworm, Spodoptera litura, and D. melanogaster [10,11].
Desmethyl-broflanilide, which has a common genesis with broflanilide, has a site of action near G3’TMD3 in the DmRDL subunit [10,35]. Nakao et al. (2013) reported that the volume of amino acid at G3’TMD3 is a factor that determines the inhibitory activity of desmethyl-broflanilide [10]. Therefore, three mutations (G3’M/S/QTMD3), which eliminate the insecticidal sensitivity with only a minor change in activity of GABAR, were selected for further assay in vivo in this study.
CRISPR/Cas9 is a powerful tool for generating specific mutations to validate gene function, e.g. insecticide resistance in D. melanogaster [36–41]. So far, several studies adopting this “reverse genetic” approach have successfully demonstrated the mode of action of various types of insecticides by modifying target receptors with compelling association between mutations and phenotypes [42–46]. The mutation of G3’MTMD3 or G3’STMD3 was successfully introduced into w1118 DmRDL and heterozygote lines were established. Meanwhile, the G3’QTMD3 was successfully introduced into the nos.Cas9 strain. Finally, nos.Cas9 bearing G3’QTMD3 in the X chromosome was replaced with the w1118 allele during crossing ensuring that the three mutant (G3’M/S/orQTMD3) strains were generated with a consistent genetic background. Unfortunately, introduction of G3’M/S/orQTMD3 resulted in lethality making it impossible to obtain the homozygous mutant strains. This may be a reason for the failure of selecting broflanilide-resistant strains of P. xylostella [14]. Similarly, occurrence of homozygous-lethal effects have hampered investigations into the modes of action of particular insecticides using the CRISPR/Cas9 strategy [45,47,48]. Therefore, the G3’MTMD3, G3’STMD3 and G3’QTMD3 were maintained in heterozygous strains, and the direct linkage between homozygous lethality and target-site mutations was proven by a complementation experiment [49].
The GFP-labeled allele can be used as a useful tool to identify the homozygote and heterozygote mutants at the embryo stage of D. melanogaster, especially when the phenotypes of the gene mutations are unknown [50,51]. Thus, heterozygous mutant strains with a GFP-labeled balancer allele enabled accurate selection of homozygous G3’TMD3 mutants by fluorescence detection. Embryos bearing any of the three homozygous mutations at RDL G3’TMD3 were able to hatch where 25% of embryos from each heterozygous strain were expected to carry homozygous mutations [52]. In line with this, the proportion of homozygous G3’MTMD3, G3’STMD3 and G3’QTMD3 embryos was 24%-27% (Fig 3B). It is worth noting that the temporal characteristics of lethality varied among the three mutants during the first three days after hatching (Fig 4A).
In this study, heterozygous lines bearing mutations at G3’TMD3 did not show significant resistance to broflanilide or fluralaner (S8 Table). This is in line with a previous study using electrophysiology, where expression of heterozygous G3’TMD3 mutant RDL in D. melanogaster Mel-2 cells did not confer resistance to demethyl-broflanilide compared with the wild-type RDL alone [15]. Therefore, these in vivo and in vitro studies indicate that heterozygous mutations at RDL G3’TMD3 do not confer resistance to meta-diamides and isoxazolines. Drosophila melanogaster homozygous for G3’MTMD3 were selected at the larval stage for bioassays due to their low mortality during the first three days after hatching. These larvae were highly resistant to broflanilide and fluralaner with LC50 > 1000 mg/kg, providing convincing evidence in vivo that meta-diamides and isoxazolines share the same mode of action and directly interact with the RDL subunit. It is also worth noting that larvae homozygous for G3’MTMD3 showed low-level resistance to avermectin, indicating that avermectin might share an overlapping but weak binding mode with fluralaner and broflanilide on the GABAR. However, the toxicity of fipronil was not affected by the homozygous G3’MTMD3 mutation, which indicated a different binding site, in accord with previous studies in vivo and in vitro [6,11,22,23].
It has been previously reported that the knock-down of RDL can affect the locomotivity of D. melanogaster [53]. In this study, our results showed that D. melanogaster larvae homozygous for G3’M/S/orQTMD3 displayed a significantly reduced crawling speed compared to wild-type w1118 (Fig 4B and 4C) suggesting that the mutation results in a potential fitness cost. Physical fitness costs caused by point mutations were also reported in other species of insect pests [43,54–58]. For example, D. melanogaster bearing the homozygous R81T in the nicotinic acetylcholine receptor β1 subunit showed an increased tolerance to neonicotinoid insecticides with a dramatic decrease in fertility, locomotivity and longevity [56].
In conclusion, the G3’TMD3 residue in RDL was identified in silico and in vitro as being important for the interaction of arthropod GABAR with broflanilide or fluralaner and its role in the sensitivity of D. melanogaster to these insecticides was verified in vivo using the CRISPR/Cas9 system. Our results showed that: 1) both broflanilide and fluralaner act on the G3’TMD3 residue of the RDL subunit; 2) a heterozygous mutation at G3’TMD3 is unlikely to lead to resistance to broflanilide and fluralaner; 3) a homozygous G3’TMD3 mutation prevented development beyond the larval stage. Our results could help systematically define the interaction of meta-diamides and isoxazolines with their molecular targets as well as further understand possible routes to resistance to these insecticides. Also, our findings may aid in the design and modification of novel insecticides, important for our continual management of agricultural and sanitary pests.
Materials and methods
Ethics statement
The use of X. laevis in the present study strictly followed the ethics of the China (GB/T 35892–2018) and Nanjing Agricultural University guidelines (https://dongwu.njau.edu.cn/info/1003/1192.htm) for the protection of animal welfare.
Chemicals
Fluralaner (purity ≥ 99%) was purified from Bravecto [59]. Fipronil (purity ≥ 96%) was provided by J & K Scientific (China) Ltd. (Beijing, CHN). Avermectin (purity ≥ 92%) was provided by Jiangsu Fengyuan Bio-Engineering Co., Ltd. (Yancheng, Jiangsu Province, CHN). Broflanilide (purity ≥ 98.67%) was provided by Mitsui Chemicals Agro, Inc. (Tokyo, JPN). Reagents and solvents used in the present study were obtained from commercial suppliers.
Insect strains and mouse mRNA
Both D. melanogaster strains w1118 and nos.Cas9 (stock #54591 at Bloomington Drosophila Stock Center) were used as the G0 generation for genome-modification. The balancer strain (w1118; TM2 Ubx130/TM6B Tb1; stock #FWB00002 at Fungene Biotech) was used for balancing. The w1118; dIRE1Δ/TM3 Sb GFP strain (stock #293 at Shanghai Institute of Biochemistry and Cell Biology) was used for replacing the balancer chromosome. The deficiency strain (w1118; Df(3L)BSC170/TM6B Tb1; stock #9561 at Bloomington Drosophila Stock Center) was used for the complementation experiment. All the flies were cultured on standard fly diet at 25°C, relative humidity of 50%-60% and a 12:12 h (L: D) photoperiod. The mouse M. musculus mRNA was kindly provided by Dr. Hui-Xing Lin (Nanjing Agricultural University).
Prediction and site-directed mutagenesis of mutation sites
An alignment of amino acid sequences of GABAR subunits from arthropods and vertebrates was constructed using MEGA 7.0 software [60] and then manually adjusted using GeneDoc (version 2.6.002) software (Pittsburgh Supercomputing Center; http://www.psc.edu/biomed/genedoc/). Thirty five amino acid residues that differed between arthropod and vertebrate sequences were selected (S3 Fig and S1 Table). The C. suppressalis RDL homology model was constructed by SWISS-MODEL (https://swissmodel.expasy.org/) using the human GABAAR-β3 homopentamer (PDB code: 4COF) as template. The constructed model was evaluated by the online programs of PROCHECK [61,62] and ProSA-web [63]. The structure of fluralaner was built using SYBYL-X 2.1 software (Tripos Inc. St. Louis, CA) running on a Windows 7 workstation, and was then docked onto the CsRDL model using Surflex-Dock module of SYBYL-X. 2.1. The potential docking site(s) were generated using the residue-based mode of Surflex-Dock with other parameters selected as default. Different single- or double-point mutations (S2 Table) of CsRDL were generated using the Mutate Monomers module of SYBYL-X 2.1 and then the mutant models were minimized under the Tripos force field with MMFF94 charges by the Powell method with a gradient convergence criterion of 0.005 kcal/mol Å. Mutant amino acid residues, which lead to significant changes in binding affinity of fluralaner, were selected for further analysis.
Heterologous expression of GABARs in X. laevis oocytes
All predicted binding sites were mutated using specific primers (S11 Table) and the Fast Mutagenesis System (TransGen Biotech, Beijing, CHN) as reported previously [64]. GABAR subunit coding sequences were cloned into the pGH19 plasmid for expression in X. laevis oocytes. Procedures for oocyte preparation and cRNA injection into oocytes were identical to those described previously [65]. The ratio of the two injected subunits in the heteromeric Mmα1β2 and Mmα1β2-M3’GTMD3 GABARs was 1: 1.
Electrophysiological assays
Two-electrode voltage-clamp electrophysiology was performed and recorded on the Axoclamp 900A Microelectrode Amplifier platform (Molecular Devices, San Jose, CA) at a holding potential of -60 mV as previously described [65,66]. The Axon Digidata 1440A Data Acquisition System (Molecular Devices) was used to record the current signals. Oocytes were placed in a recording chamber (RC-3Z, Warner Instruments, Hamden, CT) with standard oocytes saline (SOS) medium [64] at a perfusion speed of 8–10 mL/min. Electrophysiological assays were performed at 20°C. GABA was dissolved in SOS medium and applied to oocytes for 5 s at intervals of 85 s. Concentration-response curves were obtained by sequential applications of increasing concentrations of GABA. The median effective concentration (EC50) was calculated with GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA). For antagonist assays, insecticides were first dissolved in dimethyl sulfoxide (DMSO) then diluted with SOS medium to a final DMSO concentration less than 0.1% (v/v), which did not affect a response from oocytes. Following two successive control applications of the GABA (EC50), the insecticide solution was perfused alone for 85 s before GABA (EC50) was co-applied with insecticide for 5 s, and the co-application was repeated 4–5 times at 85 s intervals to obtain the maximum constant inhibition. The values of EC50, median inhibitory concentration (IC50) and the scatter plot were determined from the mean of 3–15 replicates using non-linear regression analysis on GraphPad Prism 5. Two values of EC50 or IC50 were considered significantly different if their 95% confidence intervals (CIs) did not overlap [67].
Bioassays
The bioassay method for D. melanogaster adults was based on ffrench-Constant and Roush (1991) [16] with slight modifications. Insecticide was coated internally on the inside of glass vials (diameter × height, 20 mm × 80 mm) by applying 150 μL of acetone containing various concentrations of each insecticide, and rolling the vials until the acetone evaporated. Fifteen female flies (1–3 days post-eclosion) were transferred into the vial as a replicate, and each vial was plugged with absorbent cotton (~ 0.5 g) soaked with 5% sucrose (4.0 mL). The mortality was recorded at 72 h for avermectin and at 24 h for the other insecticides after treatment. For these assays, each concentration was replicated three times and acetone treatment was used as a control. The median lethal concentration (LC50) was calculated by probit analysis using SPSS 17.0 (SPSS Inc., Chicago, IL). A χ2 test was used to assess how well the individual LC50 values agreed with the calculated linear regression lines.
For D. melanogaster larvae bioassays, insecticide was dissolved in acetone and then mixed with the fly diet (based on corn powder, white sugar, yeast and agar) to obtain a series of concentrations. A replicate consisting of fifteen newly-hatched (less than 1 h) larvae were placed individually in single chambers of a 48-well culture plate containing 0.15 g of prepared standard fly diet. After 24 h, the mortality was calculated. Each concentration was studied in triplicate and 1% (v/w) acetone treatment was used as a control. The LC50 value was calculated as for adult bioassays.
Genomic engineering strategy
The CRISPR/Cas9 system was used to generate point mutations altering the G3’TMD3 residue in the DmRDL subunit. First, a 447-bp genomic region encompassing the codon for G3’TMD3 was sequenced to identify possible SNPs. Potential CRISPR targets in the region of interest were identified using the online platform CRISPR Optimal Target Finder (http://tools.flycrispr.molbio.wisc.edu/targetFinder/). Two target sequences (gRNA1 and gRNA2, S6 Fig) without predicted off-target sites were selected for genomic editing.
To generate the G3’MTMD3 or G3’STMD3 mutations, Cas9 mRNA, gRNAs and donor plasmid were prepared and gently mixed before injection. Briefly, Cas9 mRNA was transcribed from the linearized plasmid MLM3613 (Addgene #42251) using the mMESSAGE mMACHINE T7 Transcription Kit (Ambion, Carlsbad, CA), polyadenylated with the E. coli Poly(A) polymerase Kit (New England Biolabs, MA, ENG) then purified with the RNeasy Mini Kit (QIAGEN, Duesseldorf, GER). In vitro transcriptions (IVT) of gRNAs were performed following the protocol of Bassett et al. (2013) [36] where templates were generated by annealing two DNA oligonucleotides (Dmrdl-gRNA1-F/R and Dmrdl-gRNA2-F/R, S11 Table). Then RNA was generated by IVT using the T7 RiboMAX Express Large Scale RNA Production System (Promega Corporation, Madison, WI) before being purified by phenol-chloroform extraction and isopropanol precipitation. The pBluescript SK (-) donor plasmids (pBS-G3M, pBS-G3S), which contained two ~1,000-bp homology arms flanking the targeted genomic region with certain modifications, was constructed de novo using primers (Dmrdl-G3-5-F/R, Dmrdl-G3M-3-F/R, Dmrdl-G3S-3-F/R, pBSK-s-F/R, S11 Table) with the Gibson Assembly Kit (New England Biolabs) for HDR.
To generate the G3’QTMD3 mutation, specific gRNA-expressing plasmid and donor plasmid were constructed. For the gRNA-expressing plasmid, a DNA fragment with two fused gRNA sequences was amplified using primers (pCFD4-gRNAs-F/R, S11 Table) and the pCFD4 plasmid (Addgene#49411) backbone as template. Then the modified fragment was recombined into Bbs I-digested pCFD4 vector with the ClonExpress II One Step Cloning kit (Vazyme, Nanjing, CHN). The donor plasmid pBS-G3Q was constructed using the Fast Mutagenesis System (Transgen) and the pBS-G3M plasmid as template with primers G3QDonor-F/R (S11 Table).
Purification and amplification of genomic DNA
Genomic DNA was extracted and purified from whole D. melanogaster using DNAiso Reagent (TaKaRa, Beijing, CHN). Using genomic DNA as template, PCR amplification with primer pairs (rdl335det-F/R, S11 Table) was typically performed with 2 × Rapid Taq Master Mix (Vazyme). The conditions used were 95°C for 3min, followed by 35 cycles of denaturation at 95°C for 15 s, annealing at 54°C for 15 s, and extension at 72°C for 30 s, ending with a final extension step at 72°C for 5 min.
Generation and identification of genome-modified D. melanogaster lines
To generate G3’MTMD3 and G3’STMD3 strains, embryo injection was performed according to standard protocols [41]. The mixture containing Cas9 mRNA (500 ng/ μL), gRNAs (250 ng/ μL of each) and donor plasmid (500 ng/ μL) were co-injected into embryos. The injected G0 adult flies were individually crossed to the balancer strain (w1118; TM2 Ubx130/TM6B Tb1) and analyzed for HDR alleles (S7 Fig). G1 adult flies from positive lines with a TM2 Ubx130 allele were individually crossed with the balancer strain and screened. Subsequently, the G2 adult flies with both HDR and TM2 Ubx130 alleles were self-crossed to generate heterozygous mutant strains (G3). Both of the mutant strains were verified by sequencing and maintained by self-crossing.
To generate G3’QTMD3 strains, a mixture of gRNA-expressing plasmid (200 ng/μL) and donor plasmid (150 ng/μL) were injected into embryos of nos.Cas9. The G0nos adult flies were individually crossed with nos.Cas9 flies. Subsequently, screening of G1nos flies were performed by extracting DNA from sets of ~15 individuals (adult flies) per vial. Once the HDR alleles were detected in these G1nos flies, other male flies from the same positive lines were individually crossed with females of the balancer strain (w1118; TM2 Ubx130/TM6B Tb1) (S7 Fig). When G2nos progeny emerged, male flies with a TM2 Ubx130 allele were screened after individually crossing with female balancer flies, and the lines arising from mutant G2nos male flies were retained. By this step, all nos.Cas9 in the X chromosome were replaced with w1118 background. In the G3nos generation, adult flies with both HDR and TM2 Ubx130 alleles were self-crossed to generate a mutant strain (G4nos), which was verified by genomic DNA sequencing.
Complementation experiment
To investigate if the observed lethality is genetically linked to the Rdl gene region, a complementation experiment was performed by crossing heterozygous mutant flies to the deficiency strain (w1118; Df(3L)BSC170/TM6B Tb1), which carries a deletion of a genomic region (3L:9058052; 3L:9183797) containing Rdl. For each strain, 10 crosses were set, and the genotypes of progeny were determined after emerging by the balancer-associated phenotypes.
Temporal tracking of lethality in homozygous mutations
Heterozygous mutant flies were crossed with the dIRE1Δ/TM3 Sb GFP strain to replace the balancer TM2 Ubx130 with TM3 Sb GFP. Female adults of G3’MTMD3/TM3 Sb GFP, G3’STMD3/TM3 Sb GFP or G3’QTMD3/TM3 Sb GFP strains were induced to oviposit on a juice plate for 2 h. Embryos were collected and transferred onto an agar plate (1.5%, w/w) for hatching. After 24 h, twenty homozygous mutants as one repetition were selected from newly-hatched larvae using the SMZ25 stereomicroscope (Nikon Corporation, Tokyo, JPN) and placed individually in single chambers of a 48-well culture plate containing 0.3 g standard fly diet. Then the survival rate of flies was recorded every 24 h until the seventh day. Three repetitions were performed for each genotype, and the w1118 strain was used as the control. Data were analyzed with GraphPad Prism 5 using the Log-rank test for trend and the Mantel-Cox test. Significant difference was determined by P values: ns indicates P > 0.05, * indicates P < 0.05, *** indicates P < 0.001.
Crawling assay
Larvae homozygous for G3’MTMD3, G3’STMD3 or G3’QTMD3 were selected for the crawling assay. Four larvae as one repetition were placed on an agar plate (1.5%, w/w) and movement was recorded using a stereomicroscope combined with the Mshot Digital Imaging System (Mshot Optoelectronics Technology Co., Ltd., Guangzhou, Guangdong Province, CHN) for 2 min. Recorded videos were converted to AVI format using Format Factory (Free Time, Shanghai, CHN), and analyzed with ImageJ (National Institutes of Health, Bethesda, MD) [68, 69]. Five repetitions were performed for each genotype, and the w1118 strain was used as the control. Crawling speed was analyzed with Student’s t-test using SPSS 17.0 (SPSS Inc.). Significant difference was determined by P values: *** indicates P < 0.001.
Supporting information
The content was filmed at 3.33 fps, played at 30.00 fps and 30-times original speed.
(MOV)
The year of discovery or first introduction is included.
(PDF)
(A) Structure model of an RDL subunit built by Modeller 10.3 based on a C. elegans GluCl structure (PDB ID: 3RHW). The G3’ (G335) residue in TMD3 is labeled in green. (B) Amino acid sequence alignment of TMD3 of RDL. Dm: Drosophila melanogaster, Md: Musca domestica, Lc: Lucilia cuprina, Am: Apis mellifera, Sl: Spodoptera litura, Bm: Bombyx mori. The GenBank accession numbers of the amino acid sequences are DmRDL (AAA2856), MdRDL (AB177547), LcRDL (AAB81966), AmRDL (AJE68941), SlRDL (BAW87784) and BmRDL (NP_001182630).
(PDF)
(PDF)
The subunit-specific number is given at the left for the first residue of each aligned sequence and the index numbers for positioning in TMD3 are shown at the bottom. To facilitate the alignment of GABAR subunits from different species, the nomenclature, which is used for TMD2 of insect RDL, was employed as well in this study. Therefore, the first amino acid residue preceding TMD3 is designated as “0”. Hs: Homo sapiens, Mm: Mus musculus, Cs: Chilo suppressalis, Ls: Laodelphax striatellus, Am: Apis mellifera, Dm: Drosophila melanogaster, Tu: Tetranychus urticae. The GenBank accession numbers of the amino acid sequences are Hsα1 (NP_001121120.1), Mmα1 (NP_034380.1), Hsβ2 (NP_000804.1), Mmβ2 (NP_001334243.1), Hsβ3 (NP_068712.1), Mmβ3 (NP_001033790.1), Hsγ2 (AAH74795.1), Mmγ2 (NP_032099.1), CsRDL (ASY91962.1), LsRDL (BAF31884.1), AmRDL (ANC68177.1), DmRDL (NP_523991.2) and TuRDL (BAJ41377.1).
(PDF)
(A) and (B), Representative concentration-dependent current traces of wild-type or G3’MTMD3 CsRDL induced by GABA. (C) and (D), Representative current traces of inhibition of GABA-induced currents by fluralaner applied to wild-type or G3’MTMD3 CsRDL.
(PDF)
A 2114-bp nucleotide sequence (chromosome 3L: 9163248–9165362) was used as the homology region in the donor plasmid. Exon 7 of the Rdl gene is marked with yellow rectangles. Dark blue rectangles indicate the gRNA targeted sequences while grey rectangles indicate the corresponding protospacer adjacent motif (PAM) triplets. The G3’TMD3 codon is marked with a red asterisk and bases for synonymous mutations are shown in light blue. The rdl335det-F/R primers used for sequencing target mutations is shown in purple half arrows.
(PDF)
(A) Procedure for the generation of G3’M/STMD3 mutant strains. HDR indicates a G3’MTMD3 or G3’STMD3 allele. (B) Procedure for the generation of the G3’QTMD3 strain. Background in the X chromosome is nos.Cas9, and HDR indicates a G3’QTMD3 mutant allele.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Note, the amino acids marked as yellow are the TMDs, and all the potential amino acid residues predicted to interact with fluralaner are shown in red, while the G3’TMD3 (wild-type) residue is marked in green and the M3’TMD3 (mutant) residue is marked in blue.
(PDF)
Acknowledgments
We thank Dr. Hui-Xing Lin (Nanjing Agricultural University) for providing the M. musculus mRNA.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the National Key R&D Program of China (grant number 2022YFD1400900) and the National Natural Science Foundation of China (grant number 31871995, www.nsfc.gov.cn) for CZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Savary S, Willocquet L, Pethybridge SJ, Esker P, McRoberts N, Nelson A. The global burden of pathogens and pests on major food crops. Nat Ecol Evol. 2019;3(3):430–9. Epub 2019/02/04. doi: 10.1038/s41559-018-0793-y . [DOI] [PubMed] [Google Scholar]
- 2.ffrench-Constant RH, Williamson MS, Davies TGE, Bass C. Ion channels as insecticide targets. J Neurogenet. 2016;30(3–4):163–77. Epub 2016/11/02. doi: 10.1080/01677063.2016.1229781 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casida JE, Durkin KA. Novel GABA receptor pesticide targets. Pest Biochem Physiol. 2015;121:22–30. Epub 2014/11/18. doi: 10.1016/j.pestbp.2014.11.006 . [DOI] [PubMed] [Google Scholar]
- 4.Sparks TC, Storer N, Porter A, Slater R, Nauen R. Insecticide resistance management and industry: the origins and evolution of the Insecticide Resistance Action Committee (IRAC) and the mode of action classification scheme. Pest Manag Sci. 2021;77(6):2609–19. Epub 2021/01/28. doi: 10.1002/ps.6254 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ffrench-Constant RH, Roush RT, Mortlock D, Dively GP. Isolation of dieldrin resistance from field populations of Drosophila melanogaster (Diptera: Drosophilidae). J Econ Entomol. 1990;83(5):1733–7. doi: 10.1093/jee/83.5.1733 . [DOI] [PubMed] [Google Scholar]
- 6.Nakao T, Kawase A, Kinoshita A, Abe R, Hama M, Kawahara N, et al. The A2′N mutation of the RDL gamma-aminobutyric acid receptor conferring fipronil resistance in Laodelphax striatellus (Hemiptera: Delphacidae). J Econ Entomol. 2011;104(2):646–52. doi: 10.1603/ec10391 . [DOI] [PubMed] [Google Scholar]
- 7.Gassel M, Wolf C, Noack S, Williams H, Ilg T. The novel isoxazoline ectoparasiticide fluralaner: Selective inhibition of arthropod γ-aminobutyric acid- and L-glutamate-gated chloride channels and insecticidal/acaricidal activity. Insect Biochem Mol Biol. 2014;45:111–24. Epub 2013/12/21. doi: 10.1016/j.ibmb.2013.11.009 . [DOI] [PubMed] [Google Scholar]
- 8.Rufener L, Danelli V, Bertrand D, Sager H. The novel isoxazoline ectoparasiticide lotilaner (Credelio™): a non-competitive antagonist specific to invertebrates γ-aminobutyric acid-gated chloride channels (GABACls). Parasites Vectors. 2017;10(1):530. doi: 10.1186/s13071-017-2470-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asahi M, Kobayashi M, Kagami T, Nakahira K, Furukawa Y, Ozoe Y. Fluxametamide: A novel isoxazoline insecticide that acts via distinctive antagonism of insect ligand-gated chloride channels. Pest Biochem Physiol. 2018;151:67–72. Epub 2018/02/06. doi: 10.1016/j.pestbp.2018.02.002 . [DOI] [PubMed] [Google Scholar]
- 10.Nakao T, Banba S, Nomura M, Hirase K. Meta-diamide insecticides acting on distinct sites of RDL GABA receptor from those for conventional noncompetitive antagonists. Insect Biochem Mol Biol. 2013;43(4):366–75. Epub 2013/02/14. doi: 10.1016/j.ibmb.2013.02.002 . [DOI] [PubMed] [Google Scholar]
- 11.Nakao T, Hirase K. A comparison of the modes of action of novel meta-diamide insecticides and conventional noncompetitive antagonists on the Spodoptera litura RDL GABA receptor. J Pestic Sci. 2013;38(3):123–8. doi: 10.1584/jpestics.D13-024 [DOI] [Google Scholar]
- 12.Yamato K, Nakata Y, Takashima M, Ozoe F, Asahi M, Kobayashi M, et al. Effects of intersubunit amino acid substitutions on GABA receptor sensitivity to the ectoparasiticide fluralaner. Pest Biochem Physiol. 2020;163:123–9. Epub 2019/11/06. doi: 10.1016/j.pestbp.2019.11.001 . [DOI] [PubMed] [Google Scholar]
- 13.Blythe J, Earley FGP, Piekarska-Hack K, Firth L, Bristow J, Hirst EA, et al. The mode of action of isocycloseram: A novel isoxazoline insecticide. Pest Biochem Physiol. 2022;187:105217. Epub 2022/08/25. doi: 10.1016/j.pestbp.2022.105217 . [DOI] [PubMed] [Google Scholar]
- 14.Sun X, Wei R, Li L, Zhu B, Liang P, Gao X. Resistance and fitness costs in diamondback moths after selection using broflanilide, a novel meta-diamide insecticide. Insect Sci. 2022;29(1):188–98. Epub 2021/04/16. doi: 10.1111/1744-7917.12917 . [DOI] [PubMed] [Google Scholar]
- 15.Nakao T, Banba S. Minireview: Mode of action of meta-diamide insecticides. Pest Biochem Physiol. 2015;121:39–46. Epub 2014/09/30. doi: 10.1016/j.pestbp.2014.09.010 . [DOI] [PubMed] [Google Scholar]
- 16.ffrench-Constant RH, Roush RT. Gene mapping and cross-resistance in cyclodiene insecticide-resistant Drosophila melanogaster (Mg.). Genet Res. 1991;57(1):17–21. Epub 04/14. doi: 10.1017/s0016672300028986 . [DOI] [PubMed] [Google Scholar]
- 17.Li A, Yang Y, Wu S, Li C, Wu Y. Investigation of resistance mechanisms to fipronil in diamondback moth (Lepidoptera: Plutellidae). J Econ Entomol. 2006;99(3):914–9. doi: 10.1603/0022-0493-99.3.914 . [DOI] [PubMed] [Google Scholar]
- 18.Zhao X, Ning Z, He Y, Shen J, Su J, Gao C, et al. Differential resistance and cross-resistance to three phenylpyrazole insecticides in the planthopper Nilaparvata lugens (Hemiptera: Delphacidae). J Econ Entomol. 2011;104(4):1364–8. doi: 10.1603/EC11074 . [DOI] [PubMed] [Google Scholar]
- 19.ffrench-Constant RH, Mortlock DP, Shaffer CD, Macintyre RJ, Roush RT. Molecular cloning and transformation of cyclodiene resistance in Drosophila: an invertebrate gamma-aminobutyric acid subtype A receptor locus. Proc Natl Acad Sci U S A. 1991;88(16):7209–13. doi: 10.1073/pnas.88.16.7209 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ffrench-Constant RH, Steichen JC, Rocheleau TA, Aronstein K, Roush RT. A single-amino acid substitution in a gamma-aminobutyric acid subtype A receptor locus is associated with cyclodiene insecticide resistance in Drosophila populations. Proc Natl Acad Sci U S A. 1993;90:1957–61. doi: 10.1073/pnas.90.5.1957 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ffrench-Constant RH, Rocheleau TA, Steichen JC, Chalmers AE. A point mutation in a Drosophila GABA receptor confers insecticide resistance. Nature. 1993;363:449–51. doi: 10.1038/363449a0 . [DOI] [PubMed] [Google Scholar]
- 22.Nakao T. Mechanisms of resistance to insecticides targeting RDL GABA receptors in planthoppers. Neurotoxicology. 2017;60:293–8. Epub 2016/03/18. doi: 10.1016/j.neuro.2016.03.009 . [DOI] [PubMed] [Google Scholar]
- 23.Nakao T, Hama M, Kawahara N, Hirase K. Fipronil resistance in Sogatella furcifera: Molecular cloning and functional expression of wild-type and mutant RDL GABA receptor subunits. J Pestic Sci. 2012;37(1):37–44. doi: 10.1584/jpestics.D11-018 [DOI] [Google Scholar]
- 24.Ozoe Y, Asahi M, Ozoe F, Nakahira K, Mita T. The antiparasitic isoxazoline A1443 is a potent blocker of insect ligand-gated chloride channels. Biochem Biophys Res Commun. 2010;391(1):744–9. Epub 2009/11/26. doi: 10.1016/j.bbrc.2009.11.131 . [DOI] [PubMed] [Google Scholar]
- 25.Ozoe Y, Kita T, Ozoe F, Nakao T, Sato K, Hirase K. Insecticidal 3-benzamido-N-phenylbenzamides specifically bind with high affinity to a novel allosteric site in housefly GABA receptors. Pest Biochem Physiol. 2013;107(3):285–92. Epub 2013/09/26. doi: 10.1016/j.pestbp.2013.09.005 . [DOI] [PubMed] [Google Scholar]
- 26.Ozoe Y, Ozoe F, Kita T, Rahman MM, Liu G, Hisano K, et al. Multiple Sites of Insecticidal Action in Ionotropic GABA Receptors. In: Maienfisch P, Stevenson TM, editors. Discovery and Synthesis of Crop Protection Products. 1204. Washington, DC: American Chemical Society; 2015. p. 431–46. [Google Scholar]
- 27.Nakata Y, Fuse T, Yamato K, Asahi M, Nakahira K, Ozoe F, et al. A single amino acid substitution in the third transmembrane region has opposite impacts on the selectivity of the parasiticides fluralaner and ivermectin for ligand-gated chloride channels. Mol Pharmacol. 2017;92(5):546–55. Epub 2017/09/08. doi: 10.1124/mol.117.109413 . [DOI] [PubMed] [Google Scholar]
- 28.Zhao CQ, Casida JE. Insect γ-aminobutyric acid receptors and isoxazoline insecticides: Toxicological profiles relative to the binding sites of [3H]fluralaner, [3H]-4′-ethynyl-4-n-propylbicycloorthobenzoate, and [3H]avermectin. J Agric Food Chem. 2014;62(5):1019–24. Epub 2014/01/24. doi: 10.1021/jf4050809 . [DOI] [PubMed] [Google Scholar]
- 29.Casida JE. Golden Age of RyR and GABA-R Diamide and Isoxazoline Insecticides: Common Genesis, Serendipity, Surprises, Selectivity, and Safety. Chem Res Toxicol. 2015;28(4):560–6. Epub 2015/02/26. doi: 10.1021/tx500520w . [DOI] [PubMed] [Google Scholar]
- 30.Wu S, Nomura Y, Du Y, Zhorov BS, Dong K. Molecular basis of selective resistance of the bumblebee BiNav1 sodium channel to tau-fluvalinate. Proc Natl Acad Sci U S A. 2017;114(49):12922–7. Epub 2017/11/20. doi: 10.1073/pnas.1711699114 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao CQ, Hwang SH, Buchholz BA, Carpenter TS, Lightstone FC, Yang J, et al. GABAA receptor target of tetramethylenedisulfotetramine. Proc Natl Acad Sci U S A. 2014;111(23):8607–12. Epub 2014/05/27. doi: 10.1073/pnas.1407379111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozoe Y. γ-Aminobutyrate- and glutamate-gated chloride channels as targets of insecticides. In: Cohen E, editor. Advances in Insect Physiology. 44. The Boulevard, Langford Lane, Kidlington, Oxford, OX51GB, UK: Academic Press; 2013. p. 211–86. [Google Scholar]
- 33.Kwon DH, Yoon KS, Clark JM, Lee SH. A point mutation in a glutamate-gated chloride channel confers abamectin resistance in the two-spotted spider mite, Tetranychus urticae Koch. Insect Mol Biol. 2010;19(4):583–91. Epub 2010/06/02. doi: 10.1111/j.1365-2583.2010.01017.x . [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Puinean AM, O´Reilly AO, Williamson MS, Smelt CLC, Millar NS, et al. Mutations on M3 helix of Plutella xylostella glutamate-gated chloride channel confer unequal resistance to abamectin by two different mechanisms. Insect Biochem Mol Biol. 2017;86:50–7. Epub 2017/05/31. doi: 10.1016/j.ibmb.2017.05.006 . [DOI] [PubMed] [Google Scholar]
- 35.Nakao T, Banba S. Broflanilide: A meta-diamide insecticide with a novel mode of action. Bioorg Med Chem. 2016;24(3):372–7. Epub 2015/08/28. doi: 10.1016/j.bmc.2015.08.008 . [DOI] [PubMed] [Google Scholar]
- 36.Bassett Andrew R, Tibbit C, Ponting Chris P, Liu J-L. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4(1):220–8. Epub 2013/07/01. doi: 10.1016/j.celrep.2013.06.020 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194(4):1029–35. Epub 2013/05/24. doi: 10.1534/genetics.113.152710 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci U S A. 2013;110(47):19012–7. Epub 2013/11/04. doi: 10.1073/pnas.1318481110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196(4):961–71. Epub 2014/01/29. doi: 10.1534/genetics.113.160713 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sebo ZL, Lee HB, Peng Y, Guo Y. A simplified and efficient germline-specific CRISPR/Cas9 system for Drosophila genomic engineering. Fly. 2014;8(1):52–7. Epub 2013/10/18. doi: 10.4161/fly.26828 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Port F, Chen H-M, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A. 2014;111(29):E2967–76. Epub 2014/07/07. doi: 10.1073/pnas.1405500111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Somers J, Nguyen J, Lumb C, Batterham P, Perry T. In vivo functional analysis of the Drosophila melanogaster nicotinic acetylcholine receptor Dα6 using the insecticide spinosad. Insect Biochem Mol Biol. 2015;64:116–27. Epub 2015/03/04. doi: 10.1016/j.ibmb.2015.01.018 . [DOI] [PubMed] [Google Scholar]
- 43.Douris V, Steinbach D, Panteleri R, Livadaras I, Pickett JA, Leeuwen TV, et al. Resistance mutation conserved between insects and mites unravels the benzoylurea insecticide mode of action on chitin biosynthesis. Proc Natl Acad Sci U S A. 2016;113(51):14692–7. Epub 2016/12/05. doi: 10.1073/pnas.1618258113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zimmer CT, Garrood WT, Puinean AM, Eckel-Zimmer M, Williamson MS, Davies TGE, et al. A CRISPR/Cas9 mediated point mutation in the alpha 6 subunit of the nicotinic acetylcholine receptor confers resistance to spinosad in Drosophila melanogaster. Insect Biochem Mol Biol. 2016;73:62–9. Epub 2016/04/24. doi: 10.1016/j.ibmb.2016.04.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Douris V, Papapostolou KM, Ilias A, Roditakis E, Kounadi S, Riga M, et al. Investigation of the contribution of RyR target-site mutations in diamide resistance by CRISPR/Cas9 genome modification in Drosophila. Insect Biochem Mol Biol. 2017;87:127–35. Epub 2017/06/29. doi: 10.1016/j.ibmb.2017.06.013 . [DOI] [PubMed] [Google Scholar]
- 46.Chen W, Gu X, Yang YT, Batterham P, Perry T. Dual nicotinic acetylcholine receptor subunit gene knockouts reveal limits to functional redundancy. Pest Biochem Physiol. 2022;184:105118. Epub 2022/05/11. doi: 10.1016/j.pestbp.2022.105118 . [DOI] [PubMed] [Google Scholar]
- 47.Huang JM, Rao C, Wang S, He LF, Wu SF. Multiple target-site mutations occurring in lepidopterans confer resistance to diamide insecticides. Insect Biochem Mol Biol. 2020;121:103367. Epub 2020/03/31. doi: 10.1016/j.ibmb.2020.103367 . [DOI] [PubMed] [Google Scholar]
- 48.Xue W, Mermans C, Papapostolou K-M, Lamprousi M, Christou I-K, Inak E, et al. Untangling a Gordian knot: the role of a GluCl3 I321T mutation in abamectin resistance in Tetranychus urticae. Pest Manag Sci. 2021;77(4):1581–93. Epub 2020/12/16. doi: 10.1002/ps.6215 . [DOI] [PubMed] [Google Scholar]
- 49.Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, Gresens JM, et al. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13(3):R21. doi: 10.1186/gb-2012-13-3-r21 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Li X, Zhou B. Drosophila ZnT1 is essential in the intestine for dietary zinc absorption. Biochem Biophys Res Commun. 2020;533(4):1004–11. Epub 2020/10/01. doi: 10.1016/j.bbrc.2020.09.077 . [DOI] [PubMed] [Google Scholar]
- 51.Yoo S, Nair S, Kim H-j, Kim Y, Lee C, Lee G, et al. Knock-in mutations of scarecrow, a Drosophila homolog of mammalian Nkx2.1, reveal a novel function required for development of the optic lobe in Drosophila melanogaster. Dev Biol. 2020;461(2):145–59. Epub 2020/02/13. doi: 10.1016/j.ydbio.2020.02.008 . [DOI] [PubMed] [Google Scholar]
- 52.Nelson CR, Szauter P. Cytogenetic analysis of chromosome region 89A of Drosophila melanogaster: isolation of deficiencies and mapping of Po, Aldox-1 and transposon insertions. Mol Gen Genet. 1992;235(1):11–21. doi: 10.1007/BF00286176 . [DOI] [PubMed] [Google Scholar]
- 53.Gowda SBM, Paranjpe PD, Reddy OV, Thiagarajan D, Vijayraghavan K. GABAergic inhibition of leg motoneurons is required for normal walking behavior in freely moving Drosophila. Proc Natl Acad Sci U S A. 2018;115(9):E2115–24. Epub 2018/02/13. doi: 10.1073/pnas.1713869115 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karageorgi M, Groen SC, Sumbul F, Pelaez JN, Verster KI, Aguilar JM, et al. Genome editing retraces the evolution of toxin resistance in the monarch butterfly. Nature. 2019;574:409–12. Epub 2019/10/02. doi: 10.1038/s41586-019-1610-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh S, Giesecke A, Damulewicz M, Fexova S, Mazzotta GM, Stanewsky R, et al. New Drosophila Circadian Clock Mutants Affecting Temperature Compensation Induced by Targeted Mutagenesis of Timeless. Front Physiol. 2019;10:A1442. doi: 10.3389/fphys.2019.01442 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Homem RA, Buttery B, Richardson E, Tan Y, Field LM, Williamson MS, et al. Evolutionary trade-offs of insecticide resistance—The fitness costs associated with target-site mutations in the nAChR of Drosophila melanogaster. Mol Ecol. 2020;29(14):2661–75. Epub 2020/06/22. doi: 10.1111/mec.15503 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choo A, Fung E, Chen IY, Saint R, Crisp P, Baxter SW. Precise single base substitution in the shibire gene by CRISPR/Cas9-mediated homology directed repair in Bactrocera tryoni. BMC Genet. 2020;21(2):127. doi: 10.1186/s12863-020-00934-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ly S, Strus E, Naidoo N. Genetic disruption of the putative binding site for Homer on DmGluRA reduces sleep in Drosophila. Sleep. 2019;43(1):1–7. doi: 10.1093/sleep/zsz190 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheng CW, Jia ZQ, Liu D, Wu HZ, Luo XM, Song PP, et al. Insecticidal spectrum of fluralaner to agricultural and sanitary pests. J Asia-Pac Entomol. 2017;20(4):1213–8. doi: 10.1016/j.aspen.2017.08.021 [DOI] [Google Scholar]
- 60.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4. Epub 2016/03/22. doi: 10.1093/molbev/msw054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–91. doi: 10.1107/s0021889892009944 [DOI] [Google Scholar]
- 62.Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8(4):477–86. doi: 10.1007/BF00228148 . [DOI] [PubMed] [Google Scholar]
- 63.Wiederstein M, Sippl MJ. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–10. Epub 2007/05/21. doi: 10.1093/nar/gkm290 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sheng CW, Casida JE, Durkin KA, Chen F, Han ZJ, Zhao CQ. Fiprole insecticide resistance of Laodelphax striatellus: electrophysiological and molecular docking characterization of A2′N RDL GABA receptors. Pest Manag Sci. 2018;74:2645–51. Epub 2018/07/04. doi: 10.1002/ps.5059 . [DOI] [PubMed] [Google Scholar]
- 65.Sheng CW, Jia ZQ, Ozoe Y, Huang QT, Han ZJ, Zhao CQ. Molecular cloning, spatiotemporal and functional expression of GABA receptor subunits RDL1 and RDL2 of the rice stem borer Chilo suppressalis. Insect Biochem Mol Biol. 2018;94:18–27. Epub 2018/02/05. doi: 10.1016/j.ibmb.2018.01.003 . [DOI] [PubMed] [Google Scholar]
- 66.Huang QT, Sheng CW, Jones AK, Jiang J, Tang T, Han ZJ, et al. Functional characteristics of the Lepidopteran ionotropic GABA receptor 8916 subunit interacting with the LCCH3 or the RDL subunit. J Agric Food Chem. 2021;69(39):11582–91. Epub 2021/09/23. doi: 10.1021/acs.jafc.1c00385 . [DOI] [PubMed] [Google Scholar]
- 67.Payton ME, Greenstone MH, Nathaniel S. Overlapping confidence intervals or standard error intervals: What do they mean in terms of statistical significance? J Insect Sci. 2003;3(1):1–6. Epub 2003/10/30. doi: 10.1093/jis/3.1.34 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brooks DS, Vishal K, Kawakami J, Bouyain S, Geisbrecht ER. Optimization of wrMTrck to monitor Drosophila larval locomotor activity. J Insect Physiol. 2016;93–94:11–7. Epub 2016/07/16. doi: 10.1016/j.jinsphys.2016.07.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suda K, Muraoka Y, Ortega-Yáñez A, Yoshida H, Kizu F, Hochin T, et al. Reduction of Rpd3 suppresses defects in locomotive ability and neuronal morphology induced by the knockdown of Drosophila SLC25A46 via an epigenetic pathway. Exp Cell Res. 2019;385(2):111673. Epub 2019/10/12. doi: 10.1016/j.yexcr.2019.111673 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The content was filmed at 3.33 fps, played at 30.00 fps and 30-times original speed.
(MOV)
The year of discovery or first introduction is included.
(PDF)
(A) Structure model of an RDL subunit built by Modeller 10.3 based on a C. elegans GluCl structure (PDB ID: 3RHW). The G3’ (G335) residue in TMD3 is labeled in green. (B) Amino acid sequence alignment of TMD3 of RDL. Dm: Drosophila melanogaster, Md: Musca domestica, Lc: Lucilia cuprina, Am: Apis mellifera, Sl: Spodoptera litura, Bm: Bombyx mori. The GenBank accession numbers of the amino acid sequences are DmRDL (AAA2856), MdRDL (AB177547), LcRDL (AAB81966), AmRDL (AJE68941), SlRDL (BAW87784) and BmRDL (NP_001182630).
(PDF)
(PDF)
The subunit-specific number is given at the left for the first residue of each aligned sequence and the index numbers for positioning in TMD3 are shown at the bottom. To facilitate the alignment of GABAR subunits from different species, the nomenclature, which is used for TMD2 of insect RDL, was employed as well in this study. Therefore, the first amino acid residue preceding TMD3 is designated as “0”. Hs: Homo sapiens, Mm: Mus musculus, Cs: Chilo suppressalis, Ls: Laodelphax striatellus, Am: Apis mellifera, Dm: Drosophila melanogaster, Tu: Tetranychus urticae. The GenBank accession numbers of the amino acid sequences are Hsα1 (NP_001121120.1), Mmα1 (NP_034380.1), Hsβ2 (NP_000804.1), Mmβ2 (NP_001334243.1), Hsβ3 (NP_068712.1), Mmβ3 (NP_001033790.1), Hsγ2 (AAH74795.1), Mmγ2 (NP_032099.1), CsRDL (ASY91962.1), LsRDL (BAF31884.1), AmRDL (ANC68177.1), DmRDL (NP_523991.2) and TuRDL (BAJ41377.1).
(PDF)
(A) and (B), Representative concentration-dependent current traces of wild-type or G3’MTMD3 CsRDL induced by GABA. (C) and (D), Representative current traces of inhibition of GABA-induced currents by fluralaner applied to wild-type or G3’MTMD3 CsRDL.
(PDF)
A 2114-bp nucleotide sequence (chromosome 3L: 9163248–9165362) was used as the homology region in the donor plasmid. Exon 7 of the Rdl gene is marked with yellow rectangles. Dark blue rectangles indicate the gRNA targeted sequences while grey rectangles indicate the corresponding protospacer adjacent motif (PAM) triplets. The G3’TMD3 codon is marked with a red asterisk and bases for synonymous mutations are shown in light blue. The rdl335det-F/R primers used for sequencing target mutations is shown in purple half arrows.
(PDF)
(A) Procedure for the generation of G3’M/STMD3 mutant strains. HDR indicates a G3’MTMD3 or G3’STMD3 allele. (B) Procedure for the generation of the G3’QTMD3 strain. Background in the X chromosome is nos.Cas9, and HDR indicates a G3’QTMD3 mutant allele.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Note, the amino acids marked as yellow are the TMDs, and all the potential amino acid residues predicted to interact with fluralaner are shown in red, while the G3’TMD3 (wild-type) residue is marked in green and the M3’TMD3 (mutant) residue is marked in blue.
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.