Figure 2.
Historical cohort description flowchart
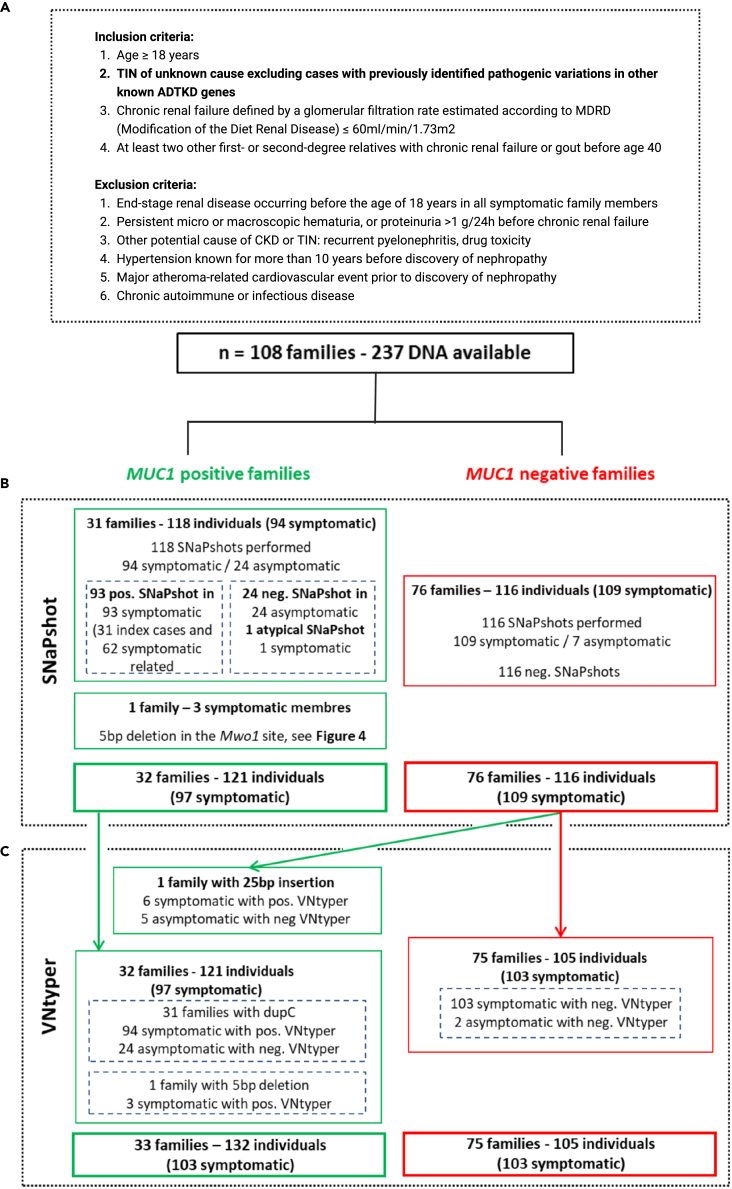
(A) The inclusion and exclusion criteria for MUC1-ADTKD are shown. In this cohort, 108 families with 237 individuals were studied by short-read sequencing. No pathogenic variant was found with standard pipeline in any gene related to ADTKD.
(B) The SNaPshot assay was performed to detect the known and recurrent MUC1 dupC variation. This method identified the dupC pathogenic variant in 31 index cases and their 62 symptomatic relatives. In one family with three affected members, our modified SNaPshot approach detected a 5bp deletion in the MwoI site.
(C) The VNtyper pipeline was applied to all individuals in the historical cohort. VNtyper re-identified all MUC1 dupC and 5bp deletion events. In one symptomatic case from MUC1-positive family (NTIH_140), linkage analysis validated the VNtyper results by confirming the segregation of the risk allele. In one family including six symptomatic members and negative SNaPshot, the pipeline detected a 25bp insertion.