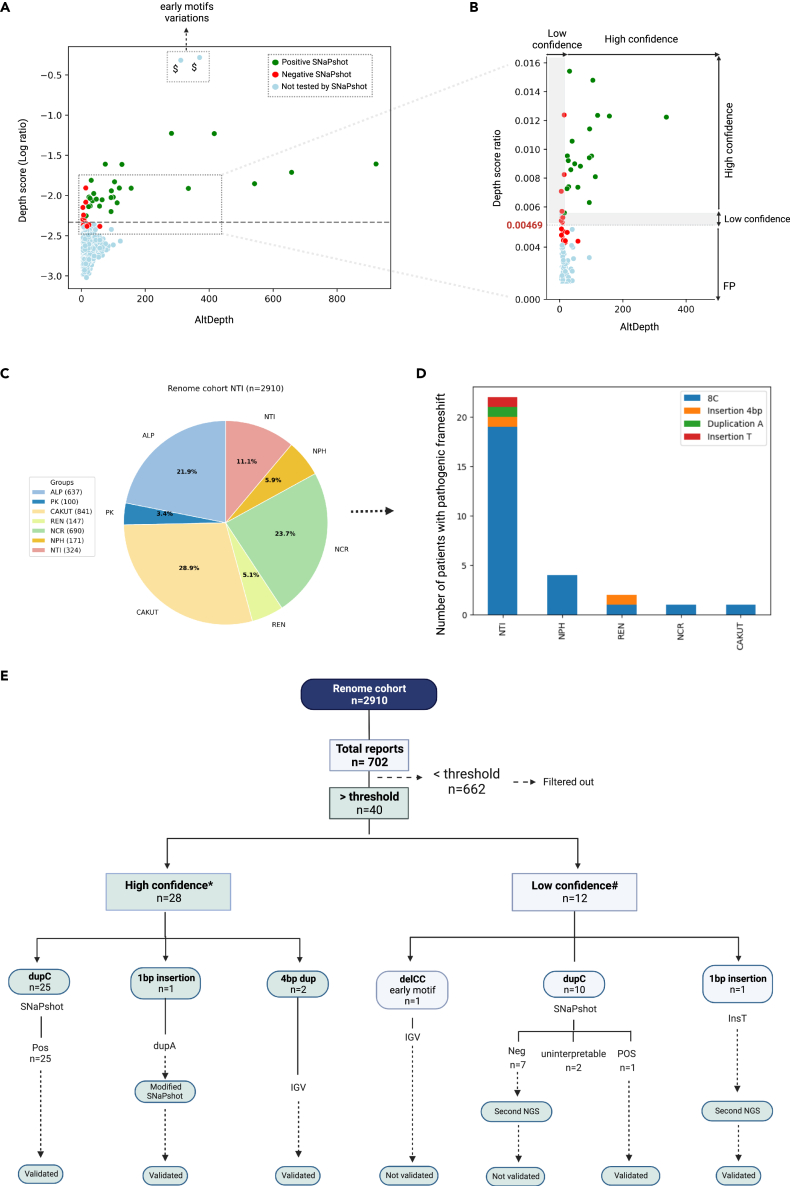
Figure 6.
Characterization of the renome cohort. VNtyper analysis of the renome cohort identified 30 MUC1-positive patients
(A and B) The depth score-adapted threshold revealed from the historical cohort was applied to the naive renome cohort. Forty cases were reported above the threshold (dashed line), and 662 instances were below the threshold. To confirm the reliability of the threshold, we analyzed all cases with Mwo1 site variations (dupC or dupA) reported above (n = 35) the threshold. We extended the SNaPshot analysis to the 11 individuals that clustered just below the threshold (i.e., between the threshold and 10% below [0.004176–0.00469]) to ensure that no true positive were missed. All cases tested below the threshold and 9/12 of the cases reported with low confidence were SNaPshot negative (red points). SNaPshot verified MUC1 dupC events in 26 cases (25 high-confidence and 1 from low-confidence cases) as well as a case with MUC1 dupA event (green points). With a second high depth NGS, we verified a patient with low confidence single base insertion as true positive. Variations identified in the early conserved motifs were validated using IGV, see Figure S5. In section B (magnification of the framed zone in A), the y axis scale was adjusted, and the plots were merged.
(C) Patients from this cohort were assigned to different groups based on the initial clinical diagnosis.
(D) MUC1 pathogenic variants were discovered in various patient groups, particularly NTI. Several cases were also identified in other cohort groups like NPH, REN, NCR, and CAKUT.
(E) The description flowchart of this cohort is shown. Note: $Individuals with variants that cannot be confirmed by SNaPshot. ∗High confidence: patients with AltDepth above 20 or depth score above 0.00515. #Low confidence: patients with AltDepth below 20 or depth score between 0.00469 and 0.00515.