Figure 1.
Generation and validation of Tet2 catalytic mutant (Tet2m/m) and knockout (Tet2−/−) ESCs
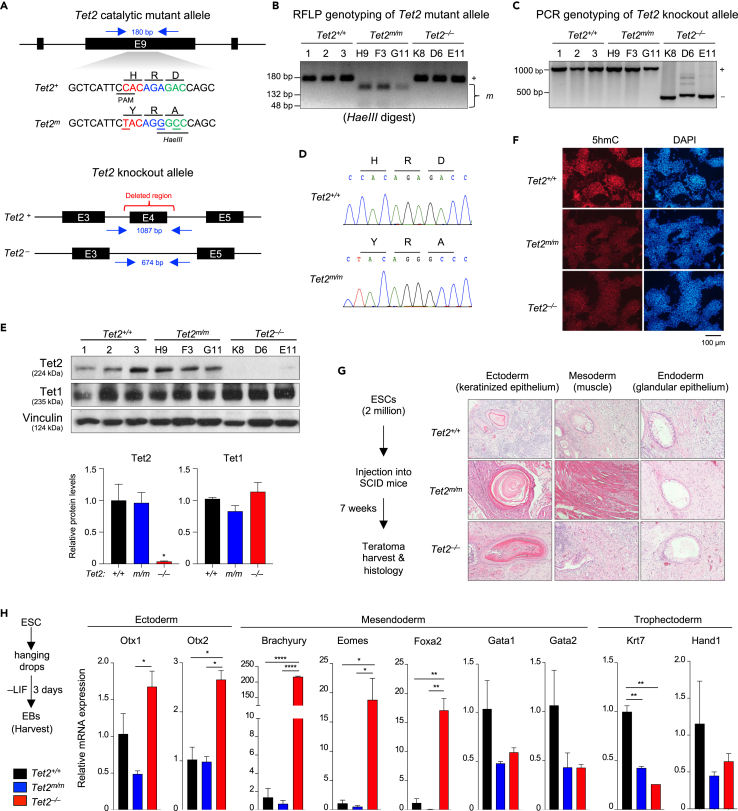
(A) Schematic of gene targeting strategy for generating Tet2m/m ESCs (top) and Tet2−/− ESCs (bottom).
(B) Validating genotypes of properly targeted Tet2m/m ESCs by RFLP (restriction fragment-length polymorphism) using HaeIII enzyme. Correctly targeted (mutated) allele bands after digestion are 132 bp + 48 bp. Allele not carrying the mutation is 180 bp. (3 independent clones were generated H9, F3, and G11).
(C) Genotyping of properly targeted Tet2−/− ESCs by PCR. Amplification of a shorter fragment (∼674 bp) confirms the correct deletion of exon 4. (3 independent clones were generated K8, D6, and E11).
(D) Sanger sequencing confirms the correct introduction of H1367Y and D1369A mutations in the Tet2 catalytic mutant allele.
(E) Quantification of Tet1 and Tet2 protein levels in ESCs of indicated genotypes by Western blot (top). Signal intensity normalized to the loading control Vinculin (average of three lines) is plotted (bottom). Note the complete loss of Tet2 protein in Tet2−/− ESCs and normal expression of catalytic mutant Tet2 in Tet2m/m ESCs. Tet1 protein levels are not changed in either genotype compared to wildtype (∗p < 0.05 versus wildtype, one-way ANOVA).
(F) Immunostaining for 5hmC in ESC of indicated genotypes using an anti-5hmC antibody. Nuclei are stained with DAPI.
(G) Hematoxylin and Eosin (H&E) staining of sections of teratomas derived from ESCs of indicated genotypes.
(H) Quantification of mRNA levels of germ layer markers in day 3 embryoid bodies (EBs) derived from wildtype, Tet2m/m and Tet2−/− ESCs by RT-qPCR. Data normalized to Gapdh. Error bars = SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.