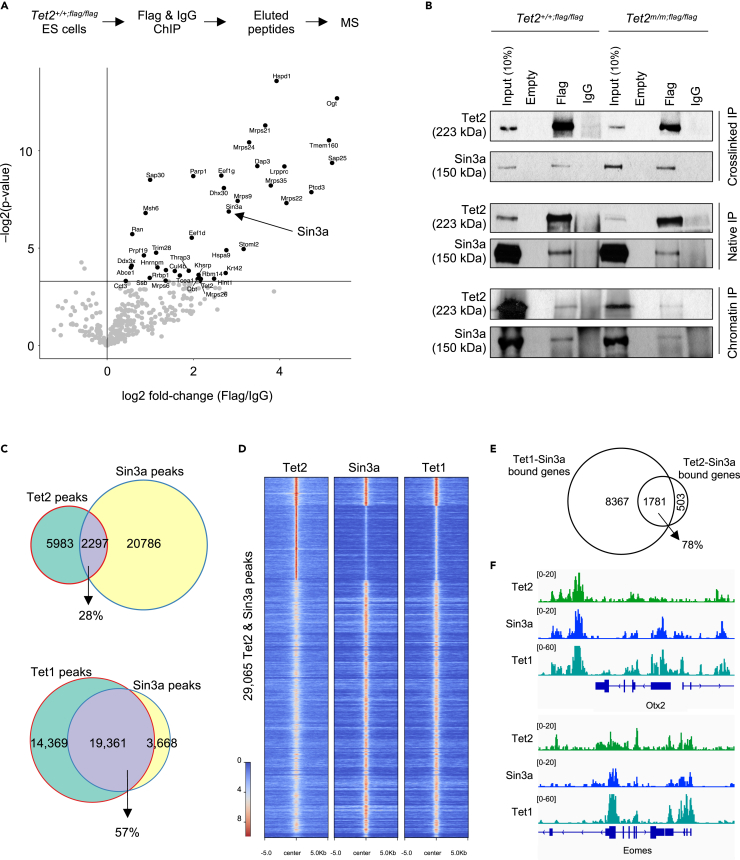
Figure 4.
Tet2 is in a complex with the histone deacetylase Sin3a in ESCs
(A) Schematic of ChIP-MS strategy for identification of endogenous binding partners of Tet2 using a Tet2flag/flag ESC line and anti-Flag antibody (top). Enrichment scores (log2 fold-change x -log2(p value)) of proteins that immunoprecipitated with Tet2 as quantified by MS is plotted (bottom). Note that several known partners of Tet2 (i.e. Ogt and Parp1) as well as new partners (i.e. Sin3a, Sap25 and Sap30) were identified.
(B) Validation of Tet2 interaction with Sin3a by immunoprecipitation (IP) of native and crosslinked nuclear lysates, and ChIP of Tet2+/+;flag/flag and Tet2m/m; flag/flag ESCs using anti-Flag antibodies and Western blot of Sin3a. Anti-IgG was used as negative control.
(C) Venn diagram showing overlap of Tet2 ChIP-seq peaks with Sin3a peaks in ESCs (top), and venn diagram showing the overlap of Tet1 and Sin3a peaks from published datasets (Chrysanthou et al., 2022) (bottom).
(D) Heatmap showing enrichment of Tet2, Sin3a, and Tet1 signal at Tet2 and Sin3a peaks (29,065 peaks total).
(E) Venn diagram showing overlap of Tet2-Sin3a co-bound genes with Tet1-Sin3a co-bound genes from our previously published datasets (Chrysanthou et al., 2022).
(F) Genome browser tracks showing the overlap of Tet2, Sin3a and Tet1 peaks at selected developmental genes.