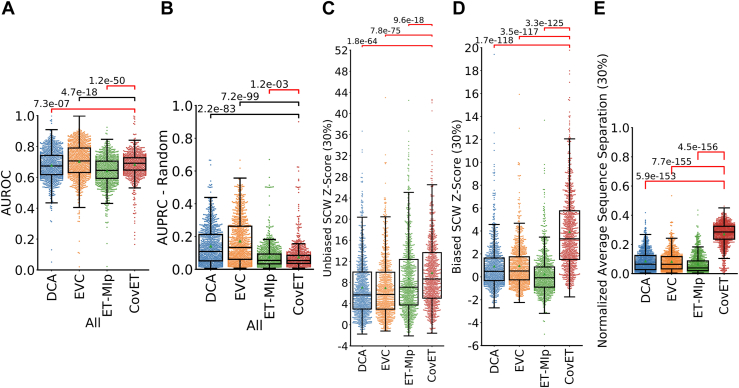
Figure 2.
CovET recapitulates the performance of existing methods in predicting structural contacts, but its predictions are more significantly clustered structurally.A, AUROC measured for all contacts (residues where Cβ are within 8 Å of each other, C⍺ for glycine) at least six residues apart. Pane (B) shows the same contacts measured at the same sequence separation categories evaluated by AUPRC. This evaluation which is less influenced by the class imbalance between contacts and noncontacts. The expected AUPRC value from a random predictor is the positive rate in the standard, which is the number of contacts/total residue pairs. AUPRC—Random evaluates whether the predictor performs better than random. C and D, unbiased (C) and biased (D) Selection cluster weighting (SCW) z-scores measured for each protein and each of the tested methods. While all methods show significant (z-score >2) clustering for most proteins, CovET shows significantly more clustering than DCA, EVCouplings (EVC), and ET-MIp in unbiased SCW z-scores and significantly better clustering than the three other methods in biased SCW z-scores. E, the average distance between pairs at 30% coverage is much higher for CovET than for any of the other methods assessed, confirming that the increase in biased SCW Z-Scores is due to clustering of residues which are significantly further apart in primary sequences. Significance was measured using the paired, two-sided Wilcoxon rank sum test. When CovET significantly outperforms other methods, the comparison bars are colored red. AUPRC, area under the precision recall curve.