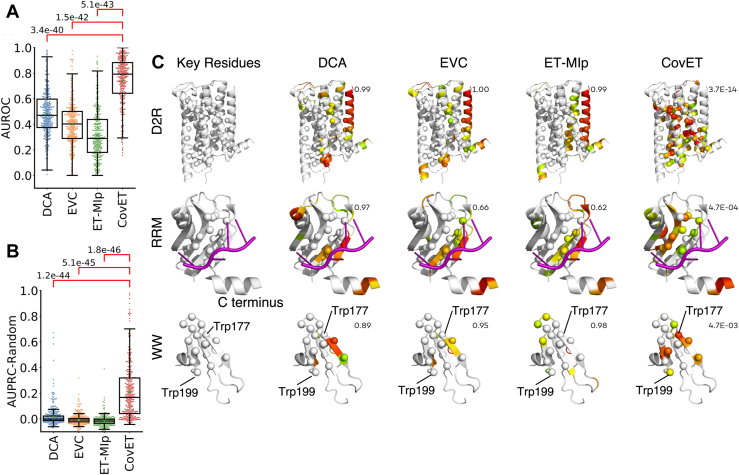
Figure 3.
Top CovET residues significantly overlap with known functional sites.A and B, CovET performs the best in recovering ligand binding sites in the pfam dataset judging by both AUROC (A) and AUPRC (B). Similar to Figure 2B, the AUPRC values are adjusted with the AUPRC of random predictors for each protein. Significance was measured using the paired, two-sided Wilcoxon rank sum test. When CovET significantly outperforms other methods, the comparison bars are colored red. C, top CovET predictions significantly overlap with known functional residues in the dopamine 2 receptor, RRM, and WW domain. Known functional residues visualized as spheres. Top 30% of residues shown on a red to green color scale, with red indicating the most highly ranked residues, and green those just at the 30% threshold. False positives shown as color scaled ribbons, false negatives shown as white spheres. Significance was measured using the one-sided hypergeometric test. The overlap between top predictions and known functional residues are evaluated with hypergeometric test. p-values are shown to the upper right of each structure. AUPRC, area under the precision recall curve; AUROC, area under the receiver operating curve; D2R, dopamine D2 receptor; RRM, RNA recognition motif.