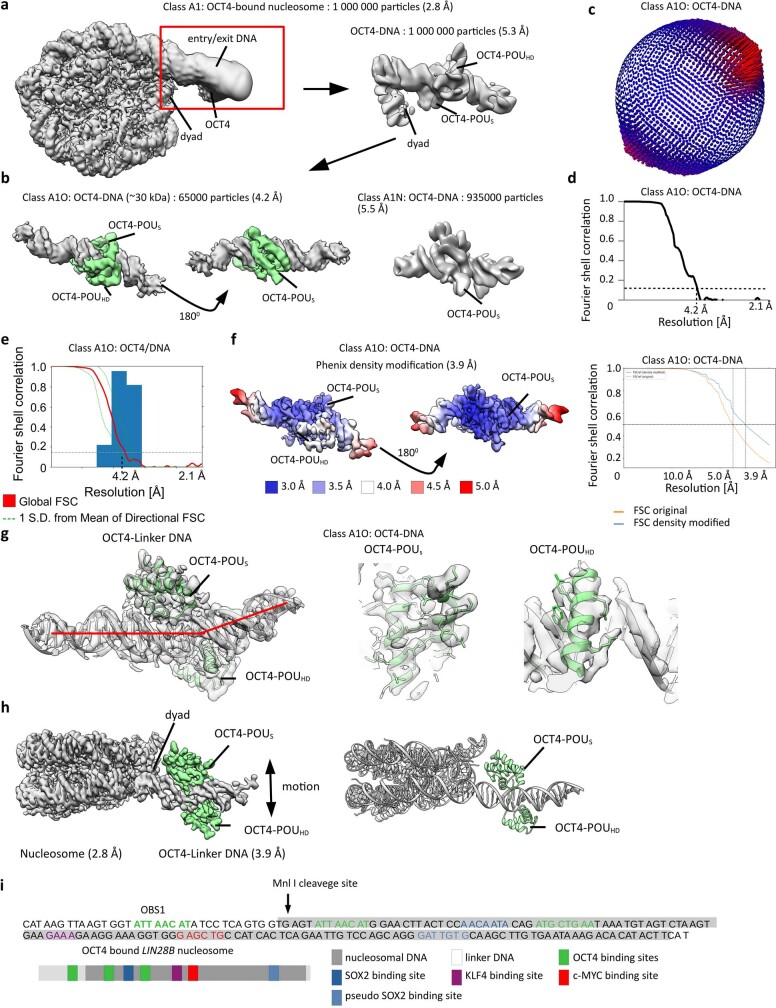
Extended Data Fig. 2. Classification of the OCT4-nucleosome complex.
a) Focused refinement of the OCT4 density from the OCT4-nucleosome (left) complex improved the resolution in the OCT4 bound region to 5.3 Å (right). b) Cryo-EM map of OCT4 region from the OCT4-nucleosome complex. Focused classification and refinements improved the resolution of this 30 kDa fragment to 4.2 Å. c) Angular distribution for OCT4. d) The fourier shell correlation (FSC) curve showing the resolution of the map. e) Directional FSC plot showing uniform resolution in all directions. f) The OCT4-DNA density from b) was modified in Phenix, which improved the resolution to 3.9 Å. The map is colored by local resolution. Fourier shell correlation (FSC) curve showing the resolution is shown on the right. g) The model of the OCT4 bound to DNA (PDB: 3L1P) was refined into the cryo-EM map. The representative region showing map quality and fit of the model is shown on the right. Red line shows the kink in the DNA. h) A composite cryo-EM map of OCT4 bound to the LIN28B nucleosome containing 182bp of DNA at 2.8-3.9 Å resolution (left). Model for the cryo-EM structure is shown on the right. i) DNA sequence and schematic representation showing LIN28B DNA positioning on the OCT4-nucleosome complex. OCT4, SOX2, KLF4 and c-MYC binding sites are labeled. The cleavage site for the restriction enzyme Mnl I (Fig. 3c) is marked with an arrow.