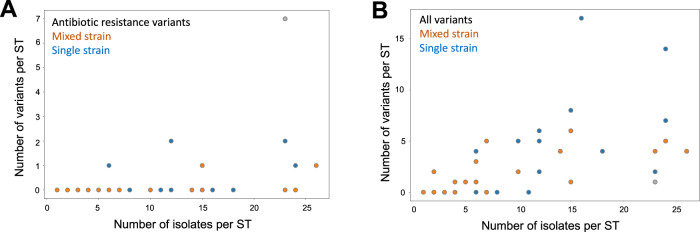
Fig. 4. Genomic drivers of resistance evolution within patients.
The abundance of variants in A genes associated with antibiotic resistance and B all variants as a function of sampling depth (i.e. number of isolates) in single strain (blue) and mixed strain (orange) patients. Each data point represents a unique combination of ST and patient. We modelled the number of variants with a negative binomial regression accounting for the infection type, number of isolates, and a possible interaction between them (n.variants ~ infection type*isolates). The number of isolates per ST was positively associated with increased number of resistance variants (0.13 +/− 0.05 variants per additional isolate; z = 2.57, p = 0.01) and other variants (0.13 +/− 0.03; z = 4.17, p < 0.001). There was no significant difference between STs from single strain and mixed strain patients in terms of A resistance variants or B genetic diversity (Supplementary Table 4a, 4b). We excluded a single strain population with an exceptionally high number of resistance variants (grey data point, see ‘Methods’), although including it did not change this conclusion (Supplementary Table 4c, 4d). The raw data is presented in the source data file.