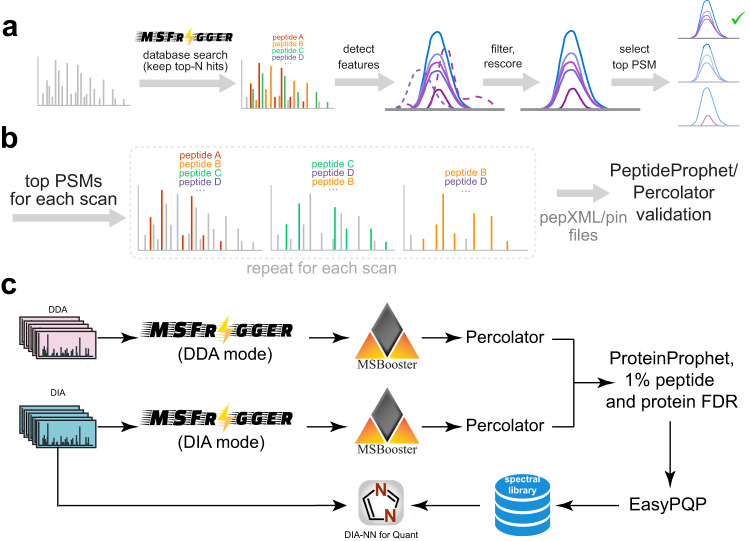
Fig. 1. Overview of MSFragger-DIA in FragPipe.
a DIA spectra are searched by MSFragger-DIA directly using precursor candidates determined from the isolation window. MSFragger-DIA builds MS1 and MS/MS spectral indexes, which are then used to detect extracted ion chromatogram (XIC) features for all fragment and precursor peaks in a peptide-spectrum match (PSM). Noisy fragment peaks are filtered out based on the XIC, PSMs are rescored, and only the top scoring PSM from each feature is kept. b Within each DIA MS/MS scan, a greedy method is used to remove matched peaks and iteratively rescore peptide candidates from the top-N list. Finally, MSFragger-DIA generates pepXML and pin files for PeptideProphet and Percolator to estimate the peptide probability in FragPipe. c Hybrid (combined DIA and DDA) data analysis workflow in FragPipe (“FP-MSF hybrid” in the main text). Both DDA and DIA data are used to build a combined spectral library. This spectral library is used to quantify peptides from the DIA data using DIA-NN.