Figure 1.
Autologous HSCT gene correction rescues the Prkdcscid phenotype but introduces on-, off-target indels and large deletions
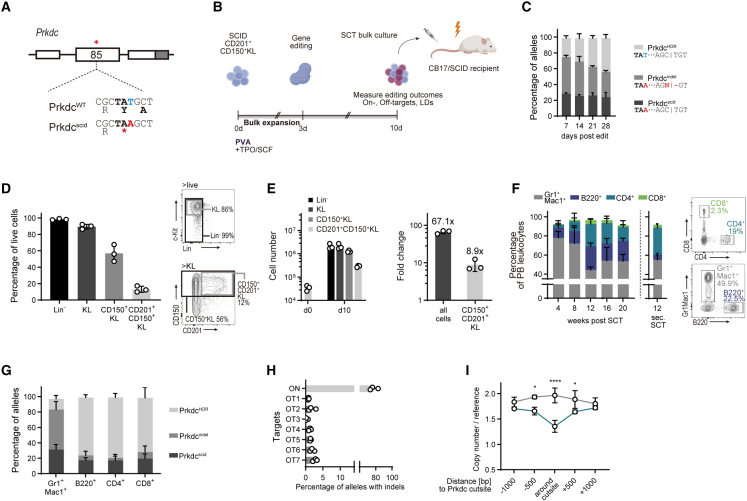
(A) Genomic context of the Prkdcscid mutation in exon 85. White boxes: exons, gray box, 3′ UTR. ∗ denotes location of Prkdcscid mutation.
(B) Experimental scheme of the gene editing and HSC expansion model.
(C) Post-editing allele distribution at the Prkdc locus, assessed by inference of CRISPR edit (ICE) (n = 3 cultures).
(D) Fractions of immunophenotypically defined HSPC populations within cultures on day 10 of culture, 7 days post-editing. Percentage of all live cells (n = 3 cultures).
(E) Absolute cell numbers (left panel) and fold-change expansion (right panel) of cultured HSPCs, day 10 of culture.
(F) Left: frequencies of peripheral blood (PB) leukocytes as percentage of all live leukocytes (n = 3 groups, 3–4 mice per group). Plot next to dashed line shows frequencies 12 weeks post-secondary SCT (n = 5 mice). Right: representative fluorescence-activated cell sorting (FACS) plots 20 weeks post-transplant.
(G) Frequencies of Prkdc alleles in sorted PB cells 20 weeks post-SCT (n = 3 experiments, 3–4 mice per group).
(H) On- and off-target (OT) activity of the Prkdc-specific gRNA, assessed with tracking of indels by decomposition (TIDE). The seven highest scoring off-target sites, as predicted by COSMID, were interrogated. See Table S1 for detailed information about the off-target sites.
(I) Copy-number analysis of Prkdc probes against reference gene (n = 3). Two-way ANOVA with Sidak’s multiple comparison test.
Error bars represent SD. ∗p < 0.05, ∗∗∗∗p < 0.0001.