Fig. 1: Overview of protocol.
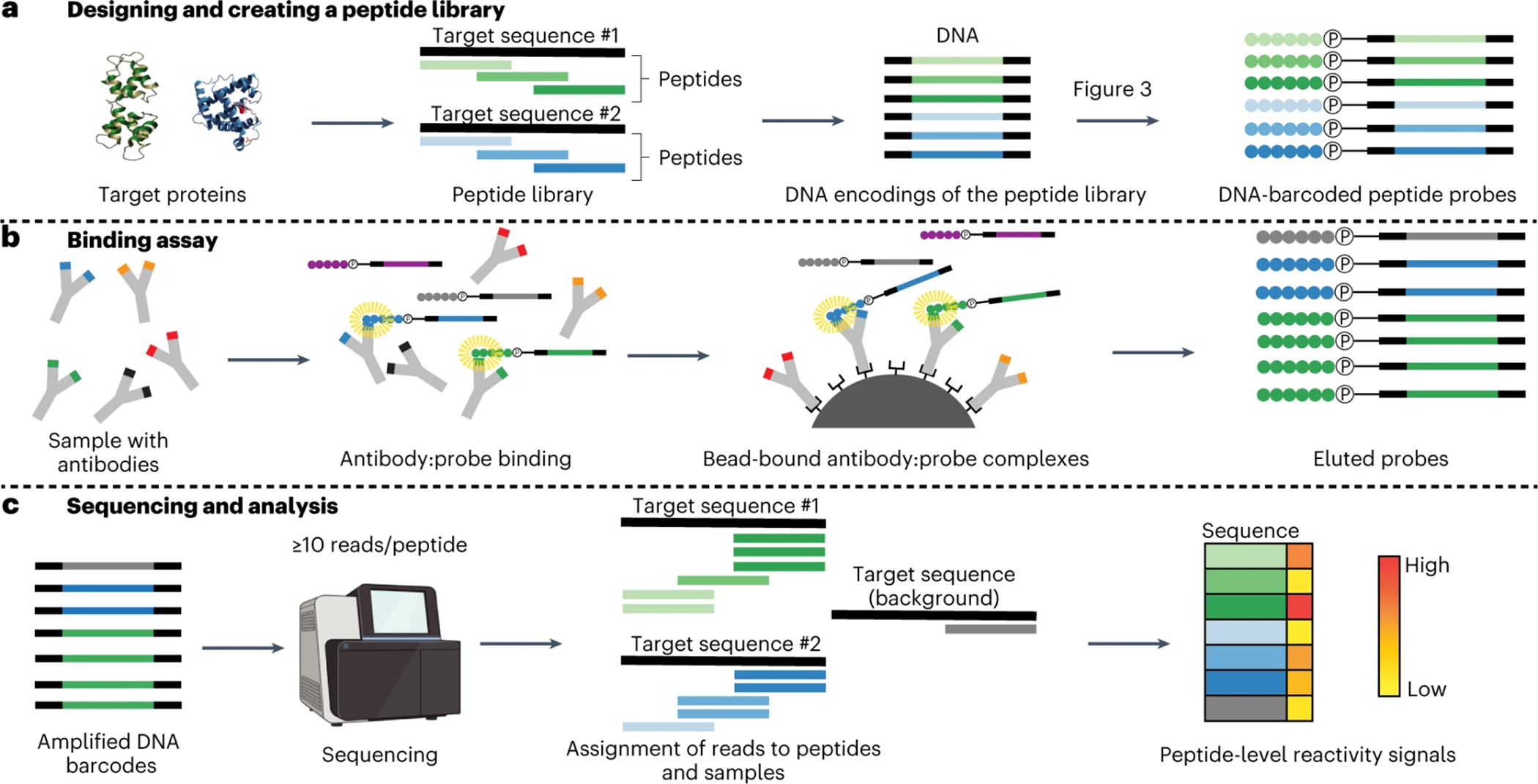
a, Design of a peptide library begins with a set of target protein sequences. An informatic approach (such as the combined sliding-window/set-cover algorithm described in the ‘Design of peptide-encoding oligonucleotides’ section of ‘Experimental design’) is then used to design a library of peptides of a user-defined length that cover the supplied target sequences (green and blue bars). Next, amino acid sequences are informatically converted into DNA encodings, and constant regions (black segments) are added to each end. The corresponding DNA oligonucleotide library is prepared by massively parallel DNA synthesis and converted in bulk to a corresponding library of DNA-barcoded peptides (‘PepSeq probes’) by using in vitro transcription and translation and taking advantage of intramolecular coupling mediated by a tethered puromycin (P)-containing molecule. b, A library of PepSeq probes, prepared as in a, are then incubated with biological samples of interest, allowing antibodies to bind their cognate epitopes (binding indicated by the yellow halos). The antibodies are then captured onto protein-bound beads via their constant regions, unbound PepSeq probes are washed away and bound PepSeq probes are eluted. c, The DNA tags of eluted PepSeq probes are amplified by PCR, and the relative abundance of each probe is quantified by using high-throughput sequencing. Changes in relative abundance provide a semi-quantitative measure of cognate antibody abundance/affinity for each peptide in the design. Created with BioRender.com.
