Fig. 2: Impact of the design algorithm and target sequence clustering approach on library design size.
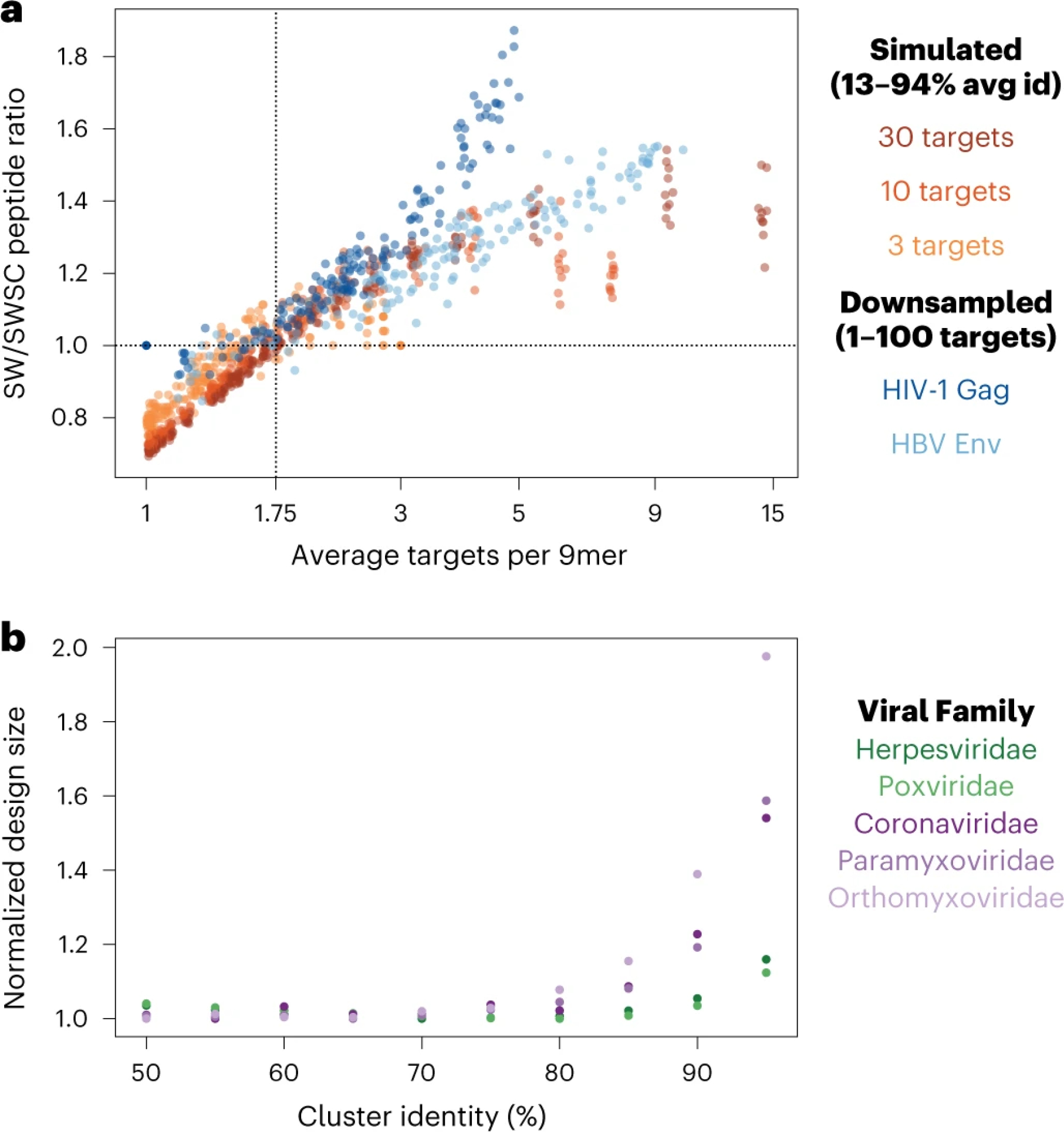
a, Comparison of performance between the sliding window only (SW) and sliding window + set cover (SWSC) design strategies. Performance is measured as the ratio of the number of 30mer peptides required to cover all unique 9mers present in the target set of proteins (points above the horizontal dotted line indicate superior performance of the SWSC algorithm). Orange points represent results from simulated datasets with a fixed number of target protein sequences (3, 10 and 30) and variable average percent identity between targets (13–94%). Blue points represent results from datasets with variable numbers (1–100) of particular viral protein sequences downsampled randomly from the sequences present in UniProt. Across all of the analyzed datasets, the SWSC design strategy more efficiently covered the target protein set when each 9mer occurred in an average of at least ~1.75 target protein sequences. b, Performance of the SWSC algorithm when target protein sequences were clustered by using Uclust at different identity thresholds (50–95%). Each color represents a different target protein dataset (37,022–1,213,326 sequences in each) generated by downloading all available protein sequences from Uniprot for five different viral families (three RNA virus families (purple) and two DNA virus families (green)). The peptide library size at each cluster identity threshold was normalized by dividing by the number of peptides contained in the smallest design for the same set of target proteins. A percent cluster identity of between 65 and 75 resulted in the smallest number of peptides needed to cover all 9mers in the selected datasets. avg, average; Env, large envelope protein; Gag, group-specific antigen protein; HBV, hepatitis B virus; id, identity.
