Fig. 3: Synthesis of DNA-barcoded peptide libraries.
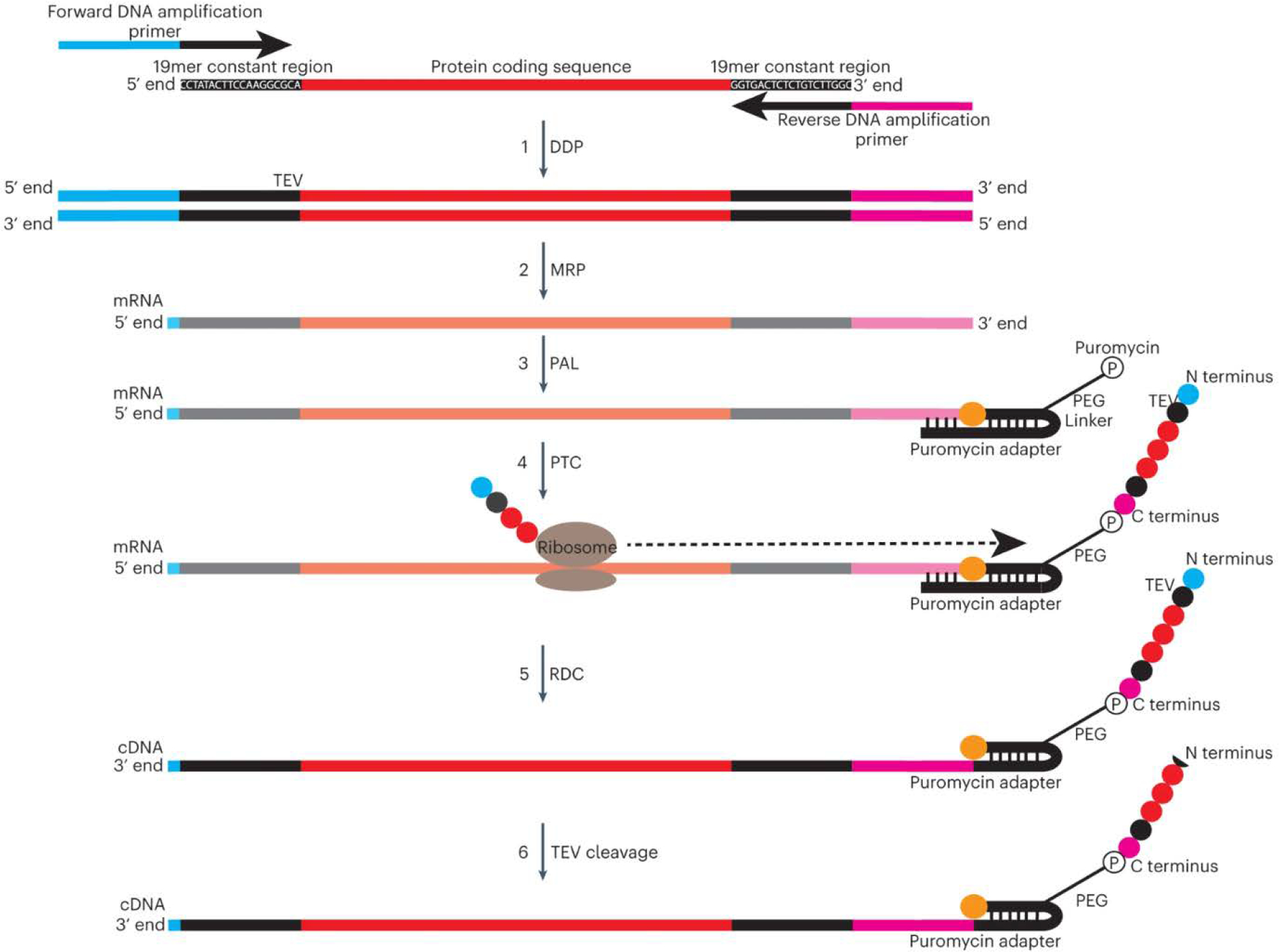
Generation of DNA-barcoded peptide (‘PepSeq’) libraries consists of the following 6 steps: 1) overlap extension PCR of a single- or double-stranded DNA library encoding the target peptides to create dsDNA templates for in vitro transcription, by using primers that anneal to flanking constant regions (dsDNA preparation, DDP); 2) in vitro transcription of dsDNA constructs to create mRNA (mRNA preparation, MRP); 3) self-splinting ligation of a puromycin-containing hairpin adapter oligonucleotide to the 3′ end of mRNA molecules (puromycin adapter ligation, PAL); 4) in vitro translation of each mRNA into its encoded peptide, and covalent intramolecular coupling of the C terminus of the translated peptide to the 3′ end of the mRNA via the puromycin molecule (peptide translatio and capture, PTC); 5) reverse transcription of the mRNA into cDNA, primed from the hairpin region of the adapter, followed by RNase digestion of the RNA strand, leaving the synthesized peptide attached to the cDNA construct (RNA to DNA conversion, RDC); and 6) tobacco etch virus (TEV) protease cleavage to remove constant-region amino acids from the N-terminal end of the peptide.
