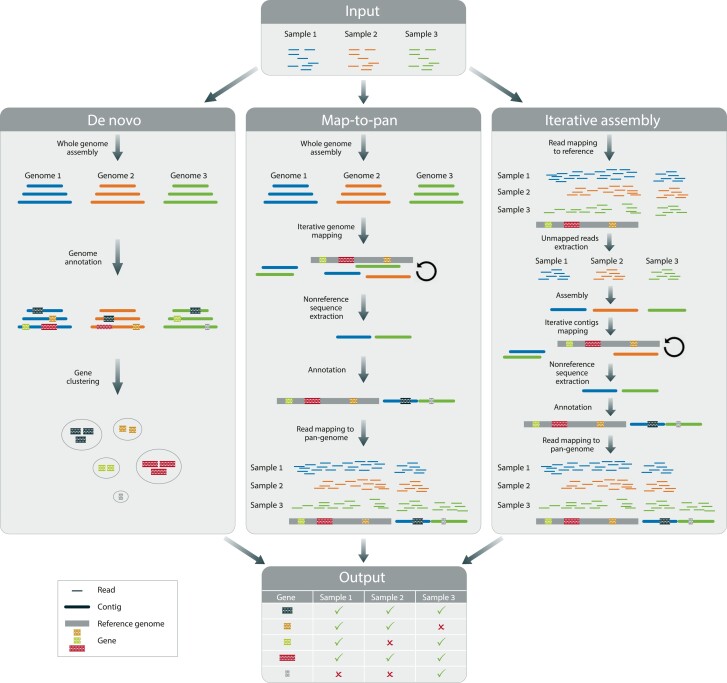
Fig. 1.
The general workflows of the DN, MTP, and IA approaches for PG construction. All approaches start from raw sequencing reads as input. The DN approach (left) begins with a whole genome assembly procedure applied to the sequencing reads of each accession, resulting in longer genomic sequences (contigs). Next, whole genome annotations of each assembled genome are performed. Gene models are detected and then clustered based on sequence similarity, with each cluster representing a pan-gene. Gene presence–absence per accession is determined based on the existence of representative genes within clusters. The MTP approach (middle) also begins with whole genome assembly but proceeds with an iterative mapping procedure to detect novel (nonreference) genomic sequences. Gene annotation is only performed on these regions, and nonreference gene models are predicted. Next, gene presence–absence is determined based on mapping of sequencing reads and analysis of gene sequence coverage in each accession. In the IA approach (right), reads are first mapped to the reference genome, and only unmapped reads are assembled into contigs which are then subject to the same steps as described for the MTP approach. All approaches result in a matrix indicating the presence or absence of each pan-gene across the accessions included in the PG.