Figure 1.
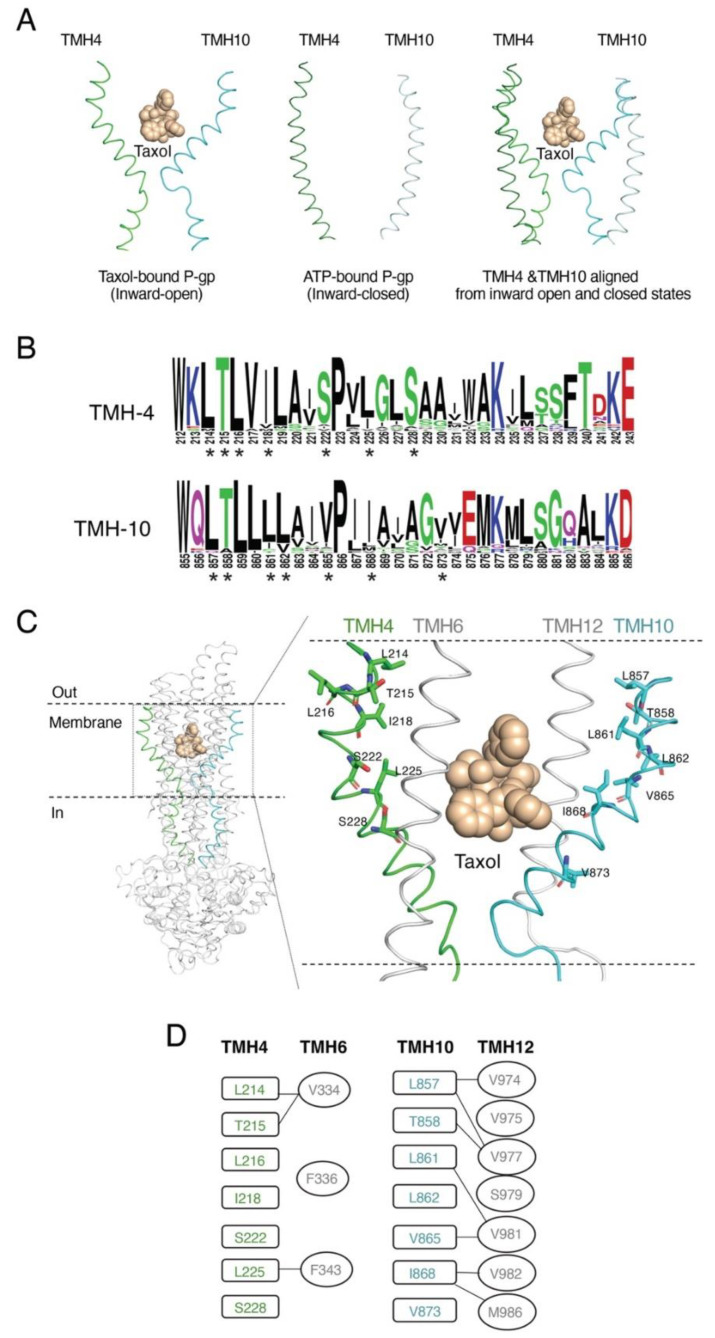
Rationale for selecting residues in TMHs 4 and 10 of P-gp. (A) Cartoon representation of TMH4 and TMH10 of human P-gp in two atomic structures. On the left, TMH4 and TMH10 are in an inward-open conformation (PDB ID: 6QEX) with Taxol in the center of the binding pocket [12]. In the middle, both helices are in the ATP-bound inward-closed conformation (PDB: 6C0V) [19]. On the right, superimposition of TMHs 4 and 10 from inward-open (green and cyan ribbon) with inward-closed (light green and light cyan) helices. (B) Sequence logo generated with the TMH4 and TMH10 sequence of P-gp from various species using the open-source software Web logo (https://weblogo.berkeley.edu/logo.cgi, accessed on 12 January 2023) The height of the residues in the logo reflects the degree of conservation. Asterisks (*) below the sequence logo indicate the conserved residues selected for mutational analysis. (C) Model of the atomic structure of P-gp based on the cryo-EM structure (PDB ID: 6QEX) with Taxol bound. In the cartoon representation of the whole P-gp molecule, TMH4 and TMH10 are highlighted in green and cyan, respectively. The enlarged area shows the residues of TMH 4 and TMH 10 selected for substitution with Ala, shown as green and cyan sticks, respectively. Both TMHs 6 and 12 are shown to indicate their proximity to TMHs 4 and 10. (D) Interactions of the TMH4 and TMH10 residues (in boxes) within 4Å of the residues (in ovals) from TMH6 and TMH12 are indicated by connecting lines.