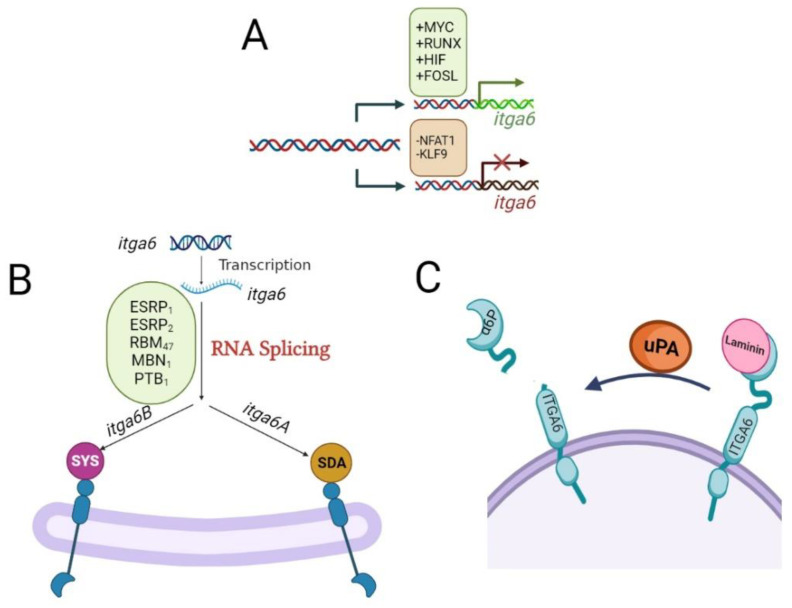
Figure 1.
Molecular mechanisms regulating the expression of itga6. (A) The expression of itga6 is regulated at multiple layers. Transcription factors, including MYC, RUNX1, HIFs, and FOSL1, bind to the itga6 promoter region and enhance its transcription, while the nuclear factor of activated T cells (NFAT1) and KLF9 suppress its transcription. (B) The pre-mRNA of itga6 undergoes alternative splicing to form two splicing variants, named itga6A (α6A) and itga6B (α6B). This alternative splicing of itga6 is regulated by epithelial splicing regulatory proteins 1 and 2 (ESRP1 and ESRP2), RNA binding motif protein 47 (RBM47), RNA-minding protein muscle blind (MBNL1), RNA-binding protein FOX2 homolog (RBFOX2), and polypyrimidine tract-binding protein 1 (PTBP1). The C-terminus of membrane-anchored proteins, such as integrins, is recognized by the PDZ domain, a structural fold found in many signal molecules. In the final three amino acids of the cytoplasmic tail of ITGA6-B, the PDZ-binding motif changes from the normal SDA sequence in ITGA6-A to an alternative atypical SYS motif. Different binding affinities of downstream effector proteins are influenced by these various sequences. (C) The proteolytic removal of the laminin-binding domain from the tumor cell surface by urokinase-type plasminogen activator (uPA; a pro-metastatic protein) facilitates the lighter molecular weight (70 kDa) of ITGA6, i.e., α6p.