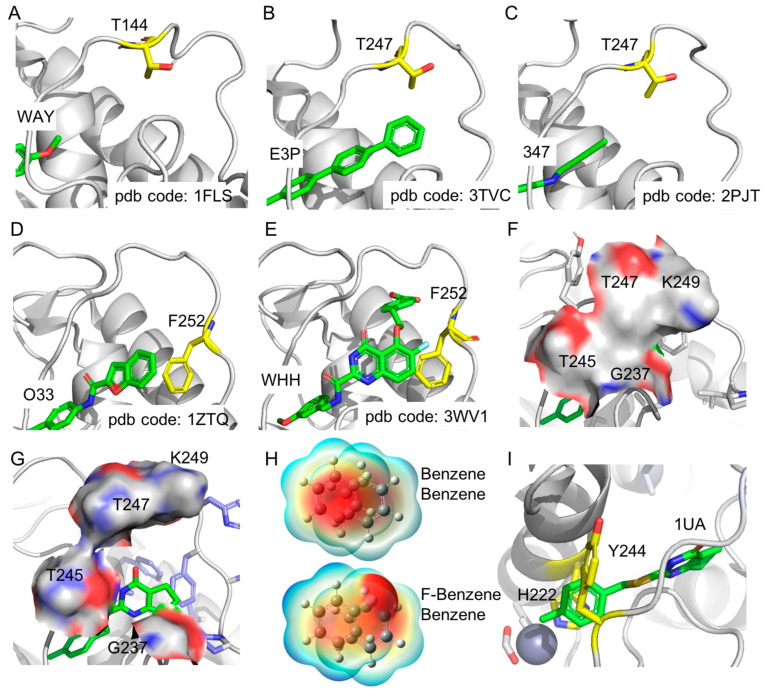
Figure 5.
The specificity loop and S1′−site of MMP−13 complexed with ligands: (A–C) The NMR structure of MMP−13 (pdb code: 1FM1) and the X-ray co-crystal structures of MMP−13 (pdb codes: 3TVC and 2PJT), respectively, which shows the flexible conformation of T144/T247 (yellow). The part of each ligand in the S1′-site is shown in green, and each ligand code is labeled; (D,E) The edge−to−face and offset π−π stacking interactions between F252 and ligands in the S1′−site of MMP−13, respectively; (F) The S1′−site closed by the hydrophobic surface made by G237, T245, T247, and K249 in the specificity loop; (G) The open state of S1′−site; (H) The electrostatic potential surface of the offset π–π stacking interaction of benzene and fluorobenzene; (I) The sandwiched π−π and π−CH (Cβ) interactions by the ligand, H222, and Y244, which could be critical for the ligand binding to MMP−13.