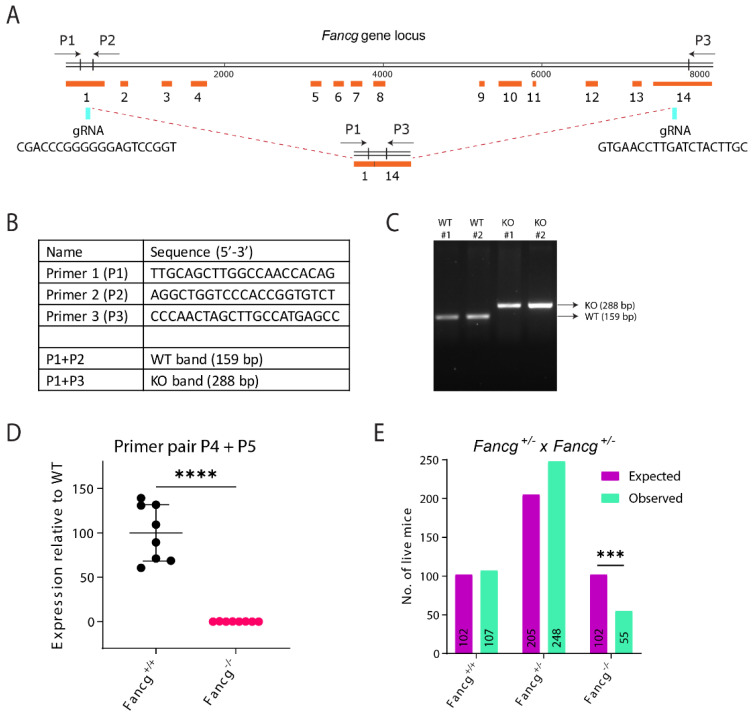
Figure 1.
Establishing a genetically and immunologically defined C57BL/6J Fancg-KO mouse model. (A) Schematic representation of the Fancg locus in the mouse and its CRISPR/Cas9 based inactivation. Cas9 activity targeted by the two gRNAs at Fancg locus in exon 1 and 14 is indicated (blue flashes). The product after the 7.5kb DNA fragment deletion is also displayed and can be detected and distinguished from wild type using the three primers P1, P2 and P3 (black arrows). (B) The table lists the sequence of primers used to screen for the knockout of Fancg allele and the primer combinations used to detect WT and Fancg-KO band simultaneously in one PCR reaction. (C) Genotype PCR distinguished WT (P1+P2) from KO(P1+P3) alleles. (D) Using qRT-PCR the lack of exons 2, 3 and 4 in the mutant cDNA of Fancg was confirmed using the primer pair id 253735709c1 from PrimerBank database. GAPDH was used as internal control and expression was plotted upon normalizing to expression of GAPDH and WT allele. p value was calculated using Welch’s t test, **** p < 0.0001. (E) Observed and expected numbers of offspring obtained from Fancg+/− intercrosses. Fancg-KO mice were born at sub-Mendelian frequencies. p values were calculated using Pearson’s Chi square test, *** p < 0.001.