Table 1.
CB1R and CB2R affinity values for compounds 13–27, 33–35, 51–55, and 56–62 in comparison with compounds 3, 63, and SR144528 a,b.
| Compd. | Structure | CB1R c Ki
f (nM) |
CB2R d Ki
f (nM) |
SI e | logKiCB2 (nM) Exp. |
logKiCB2 (nM) Pred. |
|---|---|---|---|---|---|---|
| 13 |
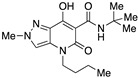
|
>10,000 | >10,000 | – | 4 | 3.19 |
| 14 |
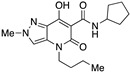
|
>10,000 | >10,000 | – | 4 | 3.53 |
| 15 |
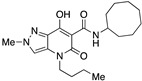
|
>10,000 | 9.03 ± 4.84 | >1107 | 1.16 | 1.6 |
| 16 |
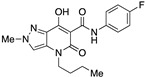
|
>10,000 | >10,000 | – | 4 | 4 |
| 17 |
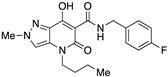
|
>10,000 | >10,000 | – | 4 | 3.73 |
| 18 |
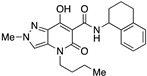
|
>10,000 | 17.31 ± 8.31 | >578 | 1.38 | 1.9 |
| 19 |
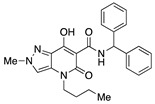
|
>10,000 | 142.25 ± 109.73 | >70 | 2.15 | 2.65 |
| 20 |
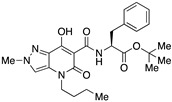
|
>10,000 | >10,000 | – | 4 | 4 |
| 21 |
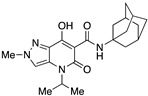
|
>10,000 | >10,000 | – | 4 | 3.59 |
| 22 |
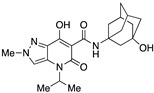
|
>10,000 | >10,000 | – | 4 | 3.39 |
| 23 |
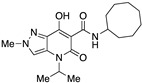
|
>10,000 | >10,000 | – | 4 | 3.05 |
| 24 |
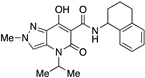
|
>10,000 | >10,000 | – | 4 | 3.43 |
| 25 |
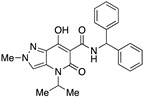
|
>10,000 | >10,000 | – | 4 | 4 |
| 26 |
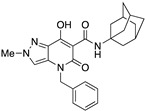
|
115.63 ± 23.66 | 0.48 ± 0.12 | 241 | −0.32 | 1 |
| 27 |
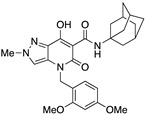
|
>10,000 | >10,000 | – | 4 | 3.52 |
| 33 |
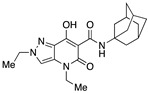
|
>10,000 | 24.04 ± 15.34 | >416 | 1.38 | 1.81 |
| 34 |
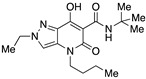
|
>10,000 | 105.85 ± 18.12 | >95 | 2.02 | 1.61 |
| 35 |
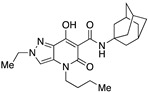
|
102.30 ± 15.23 | 0.45 ± 0.01 | 227 | −0.35 | 0.63 |
| 51 |
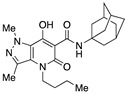
|
>10,000 | 35.61 ± 19.46 | >280 | 1.55 | 1.81 |
| 52 |
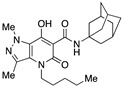
|
>10,000 | 34.10 ± 4.88 | >293 | 1.53 | 2.2 |
| 53 |
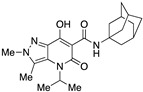
|
>10,000 | >10,000 | – | 4 | 3.88 |
| 54 |
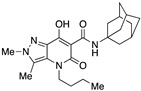
|
>10,000 | 2.16 ± 1.7 | >4630 | 0.33 | 1.2 |
| 55 |
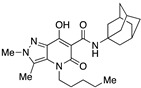
|
>10,000 | 2.11 ± 0.74 | >4739 | 0.32 | 0.69 |
| 56 |
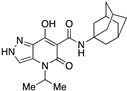
|
>10,000 | >10,000 | – | 4 | 3.54 |
| 57 |
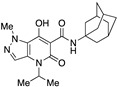
|
>10,000 | 54.31 ± 12.28 | >184 | 1.73 | 2.7 |
| 58 |
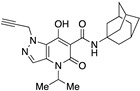
|
>10,000 | 61.76 ± 20.78 | >162 | 1.79 | 1.36 |
| 59 |
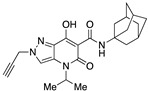
|
>10,000 | >10,000 | – | 4 | 2.8 |
| 60 |
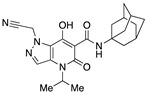
|
>10,000 | 48.46 ± 10.94 | >206 | 1.68 | 2.7 |
| 61 |
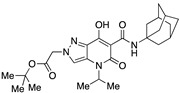
|
>10,000 | >10,000 | – | 4 | 4 |
| 62 |
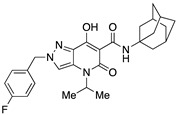
|
>10,000 | >10,000 | – | 4 | 4 |
| 63 [34] |
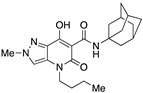
|
231 ± 76 | 2.5 ± 0.2 | 92 | – | – |
| 3 [34] |
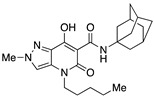
|
33.5 ± 0.9 | 0.18 ± 0.01 | 167 | – | – |
| SR144528 g,h |
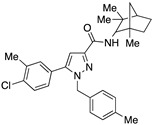
|
116 ± 22 | 1.8 ± 0.5 | 64 | – | – |
a Data represent mean values ± SD for at least three separate experiments performed in duplicate and are expressed as Ki (nM). b For both receptor binding assays, the new compounds were tested using membranes from HEK cells transfected with either CB1R or CB2R and [3H]-(–)-cis-3-[2-hydroxy-4-(1,1-dimethylheptyl)phenyl]-trans-4-(3-hydroxypropyl)cyclohexanol ([3H]CP-55,940). c CB1R: human cannabinoid type 1 receptor. d CB2R: human cannabinoid type 2 receptor. e SI: selectivity index for CB2R, calculated as Ki(CB1R)/Ki(CB2R) ratio. f Ki: inhibitor constant; that is, the concentration of the competing ligand that will bind to half the binding sites at equilibrium in the absence of radioligand or other competitors. g CB2 reference compound. h The binding affinities of reference compounds were evaluated in parallel with test compounds under the same conditions.
