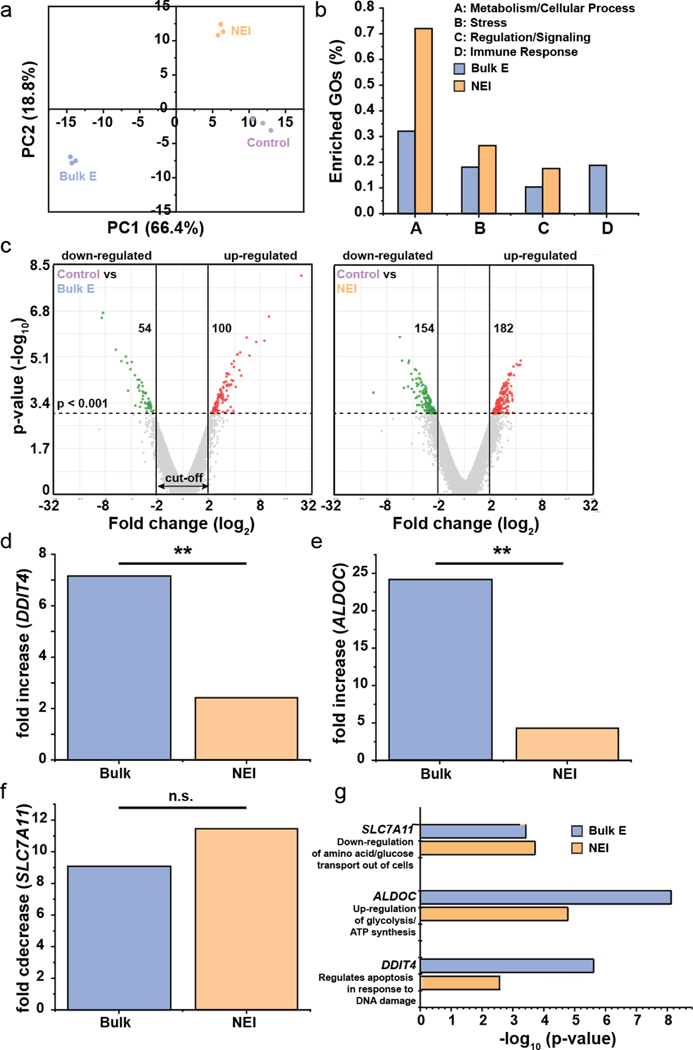
Fig. 3.
RNAseq data showing minimal gene perturbation by NEI. (a) Principal component (PC) plot showing technical replicates in each treatment group more closely resembled one another. (b) Genes in the enrich gene ontologies (GOs) which were most highly affected by treatments fell in metabolism/cellular response, stress, regulation/signaling and immune response. (c) Volcano plots showing up- and down-regulated genes after each treatment. The p-value cut off was <0.001 and log2 fold change cut-off was <−2 or >2. The number of genes that were up- or down-regulated are shown in the respective plot. (d-e) Bulk electroporation and NEI resulted in fold increase in DDIT4, a gene involved in regulation of apoptosis in response to DNA damage and ALDOC, a gene involved in glycolysis and ATP synthesis. However, bulk electroporation led to greater gene expression changes than NEI, demonstrating greater cellular perturbation by the former. (f) SLC7A11 which down-regulates the transport of amino acids and glucose out of cells was significantly down-regulated by bulk electroporation and NEI but the fold change between both treatments was not statistically significant. (g) -log10 (p-value) of DDIT4, ALDOC and SLC7A11, showing that the changes in their expressions are statistically significant from the controls. **: p < 0.001, n.s.: not significant.