Abstract
Durable T cell responses to SARS-CoV-2 antigens after infection or vaccination improve immune-mediated viral clearance. To date, population-based surveys of COVID-19 adaptive immunity have focused on testing for IgG antibodies that bind spike protein and/or neutralize the virus. Deployment of existing methods for measuring T cell immunity could provide a more complete profile of immune status, informing public health policies and interventions.
INTRODUCTION
In the early stages of the pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), public health measures in the United States were focused on minimizing spread of coronavirus disease 2019 (COVID-19) and protecting vulnerable populations through lockdowns and social distancing measures. With the emergency use authorization of multiple vaccines, the goal of achieving herd immunity appeared attainable. By May 2021, the combined vaccine- and infection-induced seroprevalence rate was reported to have reached 83.3% in the United States based on blood donor serology (1), with the caveat that these findings may not have been representative of the entire U.S. population. Although vaccinations markedly reduced both SARS-CoV-2 infections and infection-related morbidity and mortality, we have witnessed the emergence of variants with decreased susceptibility to antibody-mediated neutralization (2). In the United States, hospitals have been overwhelmed with new patients with COVID-19, and mortality rates increased during the Delta variant surge. Emergence of the Omicron variant demonstrated that waves of SARS-CoV-2 reinfection can occur, even in countries with high population immunity due to prior infection and vaccination (3). SARS-CoV-2 will continue to evolve, increasing the likelihood of evading host immunity. We must use every tool at our disposal to enable societal movement and freedom while also protecting individuals from clinically severe disease and mortality. To accomplish this goal, a comprehensive understanding of the adaptive immune response to SARS-CoV-2 infection is imperative.
Cellular immunity plays a crucial role in resolution of SARS-CoV-2 infection
The adaptive immune system is composed of two separate, complementary branches that respond to SARS-CoV-2 infection through distinct but overlapping mechanisms and with differing kinetics (4–6). COVID-19 vaccination and infection by SARS-CoV-2 induce both humoral immunity mediated by B cell–derived antibodies and cellular immunity mediated by T cells (4, 5, 7) and memory B cells (8, 9). However, much of the focus in vaccine development and immunity surveillance has been on the role of neutralizing antibodies (nAbs), with less emphasis on understanding the role of T cells, memory B cells, and non-nAbs that may confer protection via mechanisms such as opsonization and antibody-dependent cellular cytotoxicity. Mounting evidence suggests T cell contributions to the host immune response are required for early, broad, and durable protection from SARS-CoV-2, especially in the setting of new variants of concern (VOCs) (3, 6, 7, 10).
T cells can recognize a broad range of SARS-CoV-2 antigens after infection
By recognizing a broader range of viral epitopes, the T cell response may be more adept at responding to infection with evolving viral variants than antibodies. T cells can recognize linear determinants of proteins such as spike, including regions of the protein not subject to viral mutation-driven escape from antibodies. Moreover, the targets of nAbs are restricted to proteins on the viral surface, including the spike protein targeted by current SARS-CoV-2 vaccines, whereas T cell epitopes are derived from both structural and surface proteins. Furthermore, in contrast to other coronaviruses in which more than half of the T cell recognition targets the spike protein, the antigen hierarchy is more distributed across the SARS-CoV-2 proteome (10).
T cell epitopes are also shared among SARS-CoV-2 variants. On average, the spike-derived epitopes conserved at 100% amino acid sequence identity constituted 84.5 and 95.3% of the total CD4+ and CD8+ T cell epitopes, respectively (11). Similarly, there was no difference in the CD4+ and CD8+ T cell response to Delta spike peptides compared with ancestral spike peptides (12). In contrast, SARS-CoV-2 VOCs have resulted in partial evasion of humoral immunity with reduced nAb activity generated by either previous infection or vaccination (13, 14). Sera from vaccinated and convalescent individuals showed a four- to sixfold decrease in activity against the Delta variant. Furthermore, monoclonal antibodies used in a therapeutic capacity showed reduced neutralization and impaired binding to the Delta and Omicron spike protein (15, 16), and the Alpha and Beta variants are both refractory or resistant to therapeutic monoclonal antibodies directed against the spike protein (14). Modeling and in vitro analysis of the highly mutated Omicron variant has revealed considerable antibody escape (17). Despite the known loss of neutralizing capacity against the Delta, Omicron, and other VOC, vaccine efficacy in protecting from severe disease, hospitalization, and death has been only minimally to moderately affected, suggesting that durable cellular immune memory has a role in protecting against variants. In further support of these observations, 70 to 80% of the CD4+ and CD8+ T cell epitopes in the spike protein are not affected by Omicron mutations, and T cell responses appear to be largely preserved (18, 19).
T cells protect after reinfection and provide durable immunologic memory
Studies have shown that humoral and cellular immunity is retained after SARS-CoV-2 exposure (6, 20) or vaccination (21). However, unanswered questions remain regarding the level and duration of immunologic memory and its efficacy against SARS-CoV-2 reinfection. The limited understanding of the waning durability of host immune protection after SARS-CoV-2 infection and/or vaccination has contributed to implementation of immunization boosters shown to enhance both humoral and T cell responses (22). Although booster doses may enhance the magnitude and diversity of antibody responses in immunocompromised individuals with low antibody titers after their initial vaccination series, the protective role of T cells after reinfection is especially of interest given the loss of neutralizing capacity that has been demonstrated even in healthy participants both over time and in response to novel variants. Reports of association of optimal T cell responses with mild disease after primary infection (23) were supported by more recent accounts of protection conferred by SARS-CoV-2–specific memory T cells on secondary exposure (24, 25).
T cell responses have been detected in individuals who failed to seroconvert after asymptomatic or mild COVID-19 (4). Patients receiving B cell–depleting therapy exhibited a decreased antibody response while maintaining a similar T cell response compared with healthy controls after vaccination (26, 27). In addition, CD8+ T cell responses correlated with disease severity and mortality in patients with an impaired humoral response due to hematologic malignancies (28). A consensus is evolving around the significance of high levels of nAbs in mediating protection from SARS-CoV-2 infection, with a complementary role for T cells and memory B cells in preventing severe disease and hospitalization.
The durability of the T cell response is still under investigation; however, robust SARS-CoV-2–specific CD4+ and CD8+ T cell responses have been observed up to 1 year after infection and at least 6 months after vaccination (6, 20, 21). In addition, as previously shown, memory T cells to SARS-CoV-1 have been detected 17 years after exposure, suggesting that T cells may provide durable protection against severe COVID-19 disease (29).
T cell activity in response to SARS-CoV-2 may signal anticipated disease severity for patients with COVID-19
Insights gained during the pandemic point to a promising role for T cells to help us understand how SARS-CoV-2 is recognized by the adaptive immune system and how the immune response may provide insight into COVID-19 prognosis. For example, early induction of SARS-CoV-2–specific interferon-γ (IFN-γ)–secreting T cells is associated with mild disease and accelerated viral clearance (23, 30). Limited mapping of SARS-CoV-2 epitopes targeted by CD4+ and CD8+ T cells has revealed that certain viral proteins contain dominant epitopes commonly shared by multiple individuals (31). Defining these commonly shared epitopes in the context of peptide presentation by human lymphocyte antigen (HLA) alleles could lead to identification of increased or decreased COVID-19 risk in populations with particular HLA polymorphisms. A detailed analysis of B and T cell populations in patients with COVID-19 showed that more severe disease was associated with lower frequencies of CD8+ and CD4+ T cells and that there was a greater reduction of CD8+ T cells compared with CD4+ T cells in less severe disease (32). In one report, T cells unique to specific SARS-CoV-2 peptides were enriched in patients with severe disease, raising the intriguing possibility that the immune response to specific epitopes may contribute to disease severity (33). Similarly, T cells with certain phenotypes and overexuberant T cell responses have been linked to adverse outcomes (34). Defining what constitutes a protective versus harmful T cell response warrants further investigation. It will also be important to delineate the spatiotemporal dynamics and distribution of vaccine- or infection-induced T cells that correlate with better disease control, as opposed to T cell responses that reflect advanced disease and increased viral load.
Advancements in measuring T cell activation in response to SARS-CoV-2 could provide consequential insights for public health
More extensive characterization of T cell responses is needed to fully understand the immune response to SARS-CoV-2 and support the creation of informed public health strategies. The availability of laboratory technologies capable of assessing T cell responses makes the acquisition of population-level data a viable option. Antigen-specific T cell responses can be assessed by assays measuring cytokine production after antigen stimulation, such as the enzyme-linked immunosorbent spot (ELISpot) assay and intracellular cytokine staining (ICS), or the activation-induced marker (AIM) assay. ELISpot has previously been used to evaluate the duration of sustained T cell responses to SARS-CoV-2 (35). When multiplexed with flow cytometry, ICS and AIM have been used to phenotype activated cells (10, 35), allowing characterization of immunophenotypes associated with severe COVID-19. The use of tetramers has allowed for direct ex vivo characterization of SARS-CoV-2–specific T cells (36). In addition to these approaches, the U.S. Food and Drug Administration (FDA) granted emergency use authorization to a high-throughput assay that relies on next-generation sequencing (NGS) to detect and characterize SARS-CoV-2–specific T cell receptors (TCRs) for identification of prior SARS-CoV-2 infection (37). TCR immune repertoire sequencing has been applied to analyze more than 6500 samples and map epitopes to commonly shared TCR sequences (37, 38).
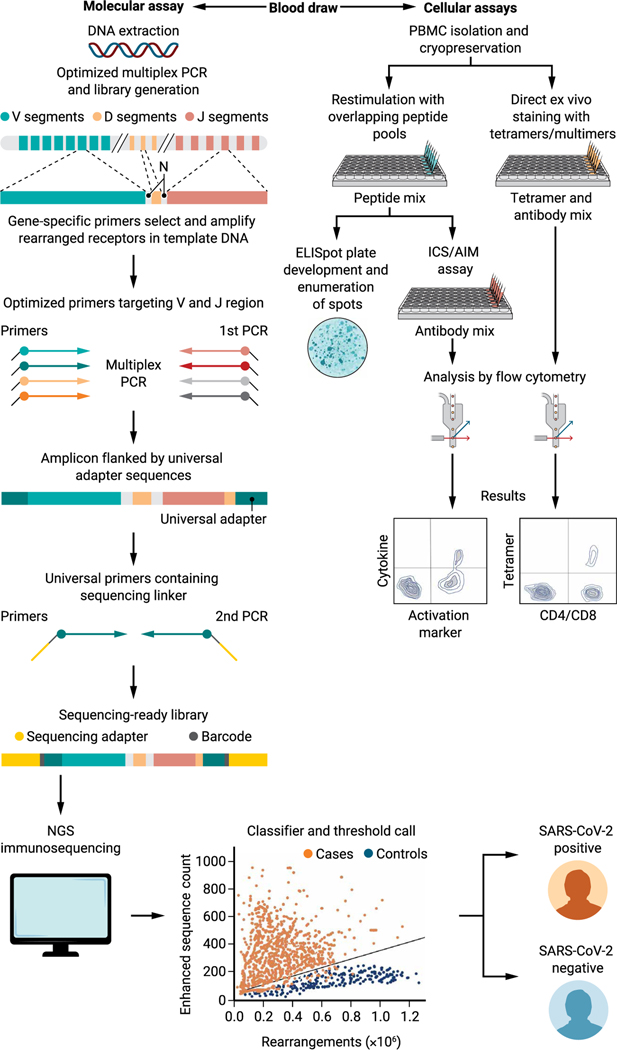
Broadly divided into molecular and cellular assays (Fig. 1), each T cell detection technology comes with inherent advantages and limitations. The choice of a specific assay to interrogate T cell responses may depend on the availability of reagents, equipment, expertise for data analysis, and the scale of the investigation, among other factors. For example, application of TCR sequencing assays based on NGS combined with trained machine learning algorithms as “classifiers” is a powerful and fast-improving area in COVID-19 diagnosis (37, 39). In the past, technical challenges and costs associated with sequencing and development of bioinformatic data analysis pipelines hindered wide-scale adoption of these assays. However, continual refinements such as minimizing amplification bias in multiplexed polymerase chain reactions (PCRs) using synthetic immune repertoires and reductions in sequencing costs are making NGS-based approaches increasingly advantageous. The requirement for DNA instead of the more logistically challenging cryopreserved, viable peripheral blood mononuclear cells (PBMCs) and the lower likelihood of NGS-based methods to be adversely affected by novel viral variants provide additional benefits in both assay accessibility and reproducibility.
Fig. 1. Schematic of molecular and cellular technologies for the evaluation of T cell responses to SARS-CoV-2.
Molecular assay workflow (left) as shown is modeled on Adaptive Biotechnologies’ immunoSEQ platform (38). Genomic DNA and a control synthetic immune repertoire template (not shown in the illustration) are PCR-amplified in parallel using primers specific for V-J sequences and subsequently for introduction of sequencing adapters and barcodes. NGS is performed, and the generated sequences are fed to the binary classifier to obtain the final results. In the classifier and threshold call graph, the enhanced sequences count (y axis) refers to the number of identified public SARS-CoV-2–associated TCRs versus the total number of unique rearrangements (x axis) within a given sample. The diagnostic model threshold is set to a high predetermined specificity against a set of holdout negative controls not used in training. Cellular assays (right) require viable PBMCs and can use either ex vivo stimulation for functional readouts, as in ELISpots and ICS/AIM assays, versus direct labeling with HLA- and peptide-specific tetramers. ICS/AIM and tetramer-based approaches involve acquisition of samples by a flow cytometer.
Most cellular assays including ELISpots and ICS/AIM coupled with flow cytometry require in vitro restimulation of T cells with HLA class I or class II peptides to measure cytokine production or, alternatively, up-regulation of surface activation markers (AIM). Assay modifications, such as the use of overlapping peptide pools, allow for coverage of all potential epitopes within a given antigenic protein and enable adoption of cellular assays for population-level assessments. ELISpots are inexpensive and the most convenient among these assays because the stimulation is followed by direct development of spots for enumeration, bypassing the need for advanced equipment and highly trained personnel. This practicality has made ELISpot an assay of choice in many clinical trials. Although ELISpot assays are easier to use, restimulation assays followed by flow cytometry–based analysis such as the ICS and AIM approaches allow for more in-depth characterization of T cells including the association of cytokines and/or activation markers with specialized T cell phenotypes including effector, memory, and other specialized subsets within the antigen-specific population. However, all in vitro restimulation assays require either fresh blood samples or healthy, cryopreserved PBMCs that retain their ability to produce cytokines during a recall response. In addition, ICS and AIM assays require relatively costly equipment and reagents, trained personnel, and are complicated by the inherent subjectivity of flow cytometry data analysis. These issues present significant barriers for the use of these assays in the clinical laboratory environment. Last, although tetramers or similar multimeric reagents confer the added advantage of binding T cells independent of their functional status, their applicability is restricted to individuals expressing specific HLA haplotypes. Considering HLA polymorphism, this approach is therefore not ideal for assessment of population-level T cell responses. Similar to many of the other cell-based assays, tetramer-based flow cytometry remains predominantly an investigational tool. A framework is needed to both understand when such tests should be translated to the clinical laboratory and what is the most appropriate regulatory framework to do so.
In the broader context, functional cellular assays based on the detection of well-established markers of T cell activation such as IFN-γ have been deployed clinically and proven useful for tuberculosis (TB), where the Mycobacterium tuberculosis–specific IFN-γ release assay (IGRA) has replaced the previous, nonspecific purified protein derivative skin test. Two IGRA tests have been approved by the FDA for TB, namely, the QuantiFERON-TB Gold In-Tube assay, which is a modified enzyme-linked immunosorbent assay, and the T-SPOT.TB (T-Spot) test, based on the principle of ELISpot (40). Research using only IGRA assays for SARS-CoV-2 infection exists (41) in the United States, including QIAGEN’s QuantiFERON SARS-CoV-2, but has not received FDA authorization for broad clinical use in COVID-19. Furthermore, the FDA announced in November 2021 that it would no longer permit any tests to be marketed clinically without first being reviewed by the FDA and being authorized under an emergency use authorization (EUA) (42).
Currently, most population-based COVID-19 surveillance studies are monitoring seroprevalence in blood donor samples, with one study analyzing over 1.4 million samples (1). The broad use of seroprevalence as a monitoring approach is likely attributable to the relative ease of doing these studies and biases them toward a focus on antibody responses. In contrast, there are no corresponding programs to analyze the T cell response at a national level. Although one goal of the U.S. Centers for Disease Control and Prevention seroprevalence surveillance program is to monitor previous exposure, multiple studies have demonstrated poor durability of antibody responses with declining titers over time (43) and, in some cases, lack of seroconversion, particularly in asymptomatic individuals (4), suggesting that data collected by seroprevalence studies may underestimate disease prevalence. Conversely, even in asymptomatic individuals, a strong and durable T cell response is detected (4), indicating that combining T cell metrics with seroprevalence may yield a more accurate measure of disease prevalence and population immunity.
Data about the cellular adaptive immune response should be expected for clinical trials supporting new therapies and vaccines. The FDA only required outcomes data (i.e., direct evidence of protection from infection and/or disease) for emergency use authorization (44). Although updated FDA guidance does recognize the use of immunogenicity studies to support efficacy of modified COVID-19 vaccine directed against SARS-CoV-2 variants and in trials in younger pediatric subjects, the agency focuses solely on measurements of antibodies. The initially announced results of the BNT162b2 vaccine trial in a pediatric population ranging from ages 2 to under 5 years were deemed insufficient to support emergency use authorization (45), yet only antibody results were considered in the analysis. This outcome failed to account for the possibility of a protective role by vaccine-induced T cells. Given the likely contribution of T cells in protection and the paucity of population-based data on the T cell response to SARS-CoV-2 infection and vaccination, future publicly funded vaccine trials should require a thorough assessment of both cellular and humoral immunity.
We believe a more complete understanding of the adaptive immune response to SARS-CoV-2 could be leveraged to inform public health policies and targeted interventions to protect vulnerable populations. In addition, comprehensive evaluation of vaccine-induced immunity will help counter vaccine misinformation that has arisen, in part, because of lack of T cell metrics. A case in point is a study that received significant media coverage, which suggested that the Ad26.COV2.S vaccine was 50% less effective against viral variants (46). These findings were directly refuted by a manufacturer-sponsored study that concluded that despite a decrease in antibody neutralization activity against viral variants, the T cell response remained unchanged, benefitting vaccine recipients with strong and persistent immunity (47).
As viral variants continue to evade host immune defenses, a second generation of vaccines or novel, tailored boosters may be necessary to enhance immunity to future SARS-CoV-2 VOC. The rapid rise in the prevalence of the heavily spike protein–mutated Omicron variant in South Africa and across the globe, as well as the Delta subvariant AY.4.2 in the United Kingdom (48), serve as an important reminder that variants will continue to emerge, resulting in decreased efficacy of the humoral response generated by previous infection with ancestral strains or vaccination. Furthermore, SARS-CoV-2 spike protein is capable of inducing host cell fusion, allowing cell-to-cell viral transmission that is refractory to antibody neutralization (49). These findings underscore the importance of cellular immunity and T cell recognition of viral variant epitopes and the cytotoxic clearance of virally infected cells. A thorough understanding of the T cell response is needed to design the next generation of vaccines that target a broad spectrum of SARS-CoV-2 antigens recognized by T cells.
As we adapt to coexist with SARS-CoV-2, it is imperative that we learn all that we can about the virus and the host immune response to drive further reductions in clinically severe disease and death. Current serology-focused testing strategies, although providing a wealth of important information, do not capture the full spectrum of immune responses to emerging variants in both healthy and vulnerable populations. Insights into the cellular immune response at the population level could be leveraged to protect against severe disease, especially in immunocompromised and susceptible individuals. Critical data supporting the role of T cells in actual protection against COVID-19 is vital and currently lacking. This issue is further evidenced by gaps in data generation regarding T cells in the development of existing and future vaccines. In addition to characterizing the role of T cells, we also need to implement clinical studies to define protective and maladaptive T cell responses and the impact they have on clinical presentations and course of disease. The ideal goal would be to define protective T cell responses after infection or vaccination or both. A recent study was conducted by Oxford Immunotec Global PLC in collaboration with Public Health England, where an assay based on the principle of ELISpot (T-SPOT Discovery SARS-CoV-2 assay) was used to measure T cell response in close to 3000 participants who were then followed up for symptomatic, PCR-confirmed SARS-CoV-2 infection (24). The study concluded that individuals with SARS-CoV-2–reactive T cells were protected from COVID-19. Because of the mutability of the virus, multiple vaccine constructs, and the diversity of the immune response, even larger-scale clinical studies may be required to define the protective T cell responses. Given the scalability and reproducibility of TCR sequencing–based approaches, this platform may be more suitable for population-level studies aimed at defining protective T cell responses. Prospective, large-cohort clinical studies that recruit thousands of volunteers may be required to find and characterize T cell correlates of protection using NGS assays, such as Adaptive Biotechnologies’ immunoSEQ (38). Serology testing can be conducted in parallel in these volunteers to gain further insights on correlation of humoral and cellular immunity. Evidence suggests that studying the cellular response to SARS-CoV-2 could allow for risk stratification among individuals as we isolate the variables contributing to increased morbidity and mortality (27). Advances in T cell–based molecular testing technologies enable evaluation of the broader adaptive immune response, can improve messaging for future vaccination campaigns, and may help inform and expedite public health strategies. Ultimately, we believe that understanding the specific and complementary roles of the adaptive immune response to SARS-CoV-2 will further decrease our vulnerability to a virus that has proven to be a formidable adversary.
Acknowledgments:
We wish to thank S. Shafiani, K. Patel, and K. MacIntosh of Adaptive Biotechnologies for medical writing support.
Funding:
Additional medical writing support was funded by Adaptive Biotechnologies and provided by L. Mitchell, R. Salmon, and M. Styers of BluPrint Oncology Concepts LLC.
Competing interests:
S.V. is a consultant or advisor for Immunai and ADC Therapeutics. L.B. declares employment, equity interest, and a leadership role at Adaptive Biotechnologies. W.G.M. has no competing interests to declare. E.J.W. is a consultant or advisor for Merck, Marengo, Janssen, Related Sciences, Synthekine, and Surface Oncology and a founder of Surface Oncology, Arsenal Biosciences, and Danger Bio. E.J.W. is an inventor on a patent (U.S. Patent number 10,370,446) submitted by Emory University that covers the use of PD-1 blockade to treat infections and cancer.
REFERENCES AND NOTES
- 1.Jones JM, Stone M, Sulaeman H, Fink RV, Dave H, Levy ME, Germanio CD, Green V, Notari E, Saa P, Biggerstaff BJ, Strauss D, Kessler D, Vassallo R, Reik R, Rossmann S, Destree M, Nguyen K-A, Sayers M, Lough C, Bougie DW, Ritter M, Latoni G, Weales B, Sime S, Gorlin J, Brown NE, Gould CV, Berney K, Benoit SJ, Miller MJ, Freeman D, Kartik D, Fry AM, Azziz-Baumgartner E, Hall AJ, Neil AM, Gundlapalli AV, Basavaraju SV, Gerber SI, Patton ME, Custer B, Williamson P, Simmons G, Thornburg MJ, Kleinman S, Stramer SL, Opsomer J, Busch MP, Estimated US infection- and vaccine-induced SARS-CoV-2 seroprevalence based on blood donations, July 2020-May 2021. JAMA 326, 1400–1409 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phillips N, The coronavirus is here to stay - Here’s what that means. Nature 590, 382–384 (2021). [DOI] [PubMed] [Google Scholar]
- 3.C Pulliam JR, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, Dushoff J, Mlisana K, Moultrie H, Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv 2021.11.11.21266068 [Preprint]. 2 December 2021. 10.1101/2021.11.11.21266068. [DOI] [PMC free article] [PubMed]
- 4.Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin J-B, Olsson A, Llewellyn-Lacey S, Kamal H, Bogdanovic G, Muschiol S, Wullimann DJ, Kammann T, Emgård J, Parrot T, Folkesson E; Karolinska COVID- Study Group, Rooyackers O, Eriksson LI, Henter J-I, Sönnerborg A, Allander T, Albert J, Nielsen M, Klingström J, Gredmark-Russ S, Björkström NK, Sandberg JK, Price DA, Ljunggren H-G, Aleman S, Buggert M, Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 183, 158–168.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rijkers G, Murk J-L, Wintermans B, van Looy B, van den Berge M, Veenemans J, Stohr J, Reusken C, van der Pol P, Reimerink J, Differences in antibody kinetics and functionality between severe and mild severe acute respiratory syndrome coronavirus 2 infections. J Infect Dis 222, 1265–1269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng C, Shi J, Fan Q, Wang Y, Huang H, Chen F, Tang G, Li Y, Li P, Li J, Cui J, Guo L, Chen S, Jiang M, Feng L, Chen L, Lei C, Ke C, Deng X, Hu F, Tang X, Li F, Protective humoral and cellular immune responses to SARS-CoV-2 persist up to 1 year after recovery. Nat. Commun. 12, 4984 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewer KJ, Barrett JR, Belij-Rammerstorfer S, Sharpe H, Makinson R, Morter R, Flaxman A, Wright D, Bellamy D, Bittaye M, Dold C, Provine NM, Aboagye J, Fowler J, Silk SE, Alderson J, Aley PK, Angus B, Berrie E, Bibi S, Cicconi P, Clutterbuck EA, Chelysheva I, Folegatti PM, Fuskova M, Green CM, Jenkin D, Kerridge S, Lawrie A, Minassian AM, Moore M, Mujadidi Y, Plested E, Poulton I, Ramasamy MN, Robinson H, Song R, Snape MD, Tarrant R, Voysey M, Watson MEE, Douglas AD, Hill AVS, Gilbert SC, Pollard AJ, Lambe T; Oxford COVID Vaccine Trial Group, T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med. 27, 270–278 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, Grifoni A, Ramirez SI, Haupt S, Frazier A, Nakao C, Rayaprolu V, Rawlings SA, Peters B, Krammer F, Simon V, Saphire EO, Smith DM, Weiskopf D, Sette A, Crotty S, Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371, eabf4063 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciabattini A, Pastore G, Fiorino F, Polvere J, Lucchesi S, Pettini E, Auddino S, Rancan I, Durante M, Miscia M, Rossetti B, Fabbiani M, Montagnani F, Medaglini D, Evidence of SARS-CoV-2-specific memory B cells six months after vaccination with the BNT162b2 mRNA vaccine. Front. Immunol. 12, 740708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A, Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489–1501.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarke A, Sidney J, Methot N, Yu ED, Zhang Y, Dan JM, Goodwin B, Rubiro P, Sutherland A, Wang E, Frazier A, Ramirez SI, Rawlings SA, Smith DM, da Silva Antunes R, Peters B, Scheuermann RH, Weiskopf D, Crotty S, Grifoni A, Sette A, Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2, 100355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jordan SC, Shin B-H, Gadsden T-AM, Chu M, Petrosyan A, Le CN, Zabner R, Oft J, Pedraza I, Cheng S, Vo A, Ammerman N, Plummer J, Ge S, Froch M, Berg A, Toyoda M, Zhang R, T cell immune responses to SARS-CoV-2 and variants of concern (alpha and delta) in infected and vaccinated individuals. Cell. Mol. Immunol. 18, 2554–2556 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Beltran WF, Lam EC, Denis KS, Nitido AD, Garcia ZH, Hauser BM, Feldman J, Pavlovic MN, Gregory DJ, Poznansky MC, Sigal A, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB, Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184, 2372–2383.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, Graham BS, Mascola JR, Chang JY, Yin MT, Sobieszczyk M, Kyratsous CA, Shapiro L, Sheng Z, Huang Y, Ho DD, Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. bioRxiv 2021.01.25.428137 [Preprint]. 12 February 2021. 10.1101/2021.01.25.428137. [DOI] [PubMed]
- 15.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Guen JL, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Péré H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Lorière E, Rey FA, Schwartz O, Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596, 276–280 (2021). [DOI] [PubMed] [Google Scholar]
- 16.McCallum M, Czudnochowski N, Rosen LE, Zepeda SK, Bowen JE, Dillen JR, Powell AE, Croll TI, Nix J, Virgin HW, Corti D, Snell G, Veesler D, Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. bioRxiv 2021.12.28.474380 [Preprint]. 31 December 2021. 10.1101/2021.12.28.474380. [DOI]
- 17.Cele S, Jackson L, Khan K, Khoury D, Moyo-Gwete T, Tegally H, Scheepers C, Amoako D, Karim F, Bernstein M, Lustig G, Archary D, Smith M, Ganga Y, Jule Z, Reedoy K, San JE, Hwa S-H, Giandhari J, Blackburn JM, Gosnell BI, Karim SA, Hanekom W, Anne von Gottberg J Bhiman, Lessells RJ, Moosa M-YS, Davenport M, de Oliveira T, Moore PL, Sigal A, SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv 2021.12.08.21267417 [Preprint]. 9 December 2021. 10.1101/2021.12.08.21267417. [DOI]
- 18.Keeton R, Tincho MB, Ngomti A, Baguma R, Benede N, Suzuki A, Khan K, Cele S, Bernstein M, Karim F, Madzorera SV, Moyo-Gwete T, Mennen M, Skelem S, Adriaanse M, Mutithu D, Aremu O, Stek C, du Bruyn E, Van Der Mescht MA, de Beer Z, de Villiers TR, Bodenstein A, van den Berg G, Mendes A, Strydom A, Venter M, Giandhari J, Naidoo Y, Pillay S, Tegally H, Grifoni A, Weiskopf D, Sette A, Wilkinson RJ, de Oliveira T, Bekker L-G, Gray G, Ueckermann V, Rossouw T, Boswell MT, Bhiman JN, Moore PL, Sigal A, Ntusi NAB, Burgers WA, Riou C, T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature 603, 488–492 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfizer, Pfizer and BioNTech provide update on Omicron variant, press release (8 December 2021); www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-provide-update-omicron-variant.
- 20.Hou H, Zhang Y, Tang G, Luo Y, Liu W, Cheng C, Jiang Y, Xiong Z, Wu S, Sun Z, Xu S, Fan X, Wang F, Immunologic memory to SARS-CoV-2 in convalescent COVID-19 patients at 1 year postinfection. J. Allergy Clin. Immunol. 148, 1481–1492.e2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goel RR, Painter MM, Apostolidis SA, Mathew D, Meng W, Rosenfeld AM, Lundgreen KA, Reynaldi A, Khoury DS, Pattekar A, Gouma S, Kuri-Cervantes L, Hicks P, Dysinger S, Hicks A, Sharma H, Herring S, Korte S, Baxter AE, Oldridge DA, Giles JR, Weirick ME, McAllister CM, Awofolaju M, Tanenbaum N, Drapeau EM, Dougherty J, Long S, D’Andrea K, Hamilton JT, Laughlin MM, Williams JC, Adamski S, Kuthuru O; UPenn COVID Processing Unit, Frank I, Betts MR, Vella LA, Grifoni A, Weiskopf D, Sette A, Hensley SE, Davenport MP, Bates P, Prak ETL, Greenplate AR, Wherry EJ, mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science 374, abm0829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munro APS, Janani L, Cornelius V, Aley PK, Babbage G, Baxter D, Bula M, Cathie K, Chatterjee K, Dodd K, Enever Y, Gokani K, Goodman AL, Green CA, Harndahl L, Haughney J, Hicks A, van der Klaauw AA, Kwok J, Lambe T, Libri V, Llewelyn MJ, McGregor AC, Minassian AM, Moore P, Mughal M, Mujadidi YF, Murira J, Osanlou O, Osanlou R, Owens DR, Pacurar M, Palfreeman A, Pan D, Rampling T, Regan K, Saich S, Salkeld J, Saralaya D, Sharma S, Sheridan R, Sturdy A, Thomson EC, Todd S, Twelves C, Read RC, Charlton S, Hallis B, Ramsay M, Andrews N, Nguyen-Van-Tam JS, Snape MD, Liu X, Faust SN; COV-BOOST study group, Safety and immunogenicity of seven COVID-19 vaccines as a third dose (booster) following two doses of ChAdOx1 nCov-19 or BNT162b2 in the UK (COV-BOOST): A blinded, multicentre, randomised, controlled, phase 2 trial. The Lancet 398, 2258–2276 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan AT, Linster M, Tan CW, Bert NL, Chia WN, Kunasegaran K, Zhuang Y, Tham CYL, Chia A, Smith GJD, Young B, Kalimuddin S, Low JGH, Lye D, Wang L-F, Bertoletti A, Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 34, 108728 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wyllie D, Jones HE, Mulchandani R, Trickey A, Taylor-Phillips S, Brooks T, Charlett A, Ades AE; EDSAB-HOME investigators, Moore P, Boyes J, Hormis A, Todd N, Reckless I, Makin A, Oliver I, SARS-CoV-2 responsive T cell numbers and anti-Spike IgG levels are both associated with protection from COVID-19: A prospective cohort study in keyworkers. medRxiv 2020.11.02.20222778 [Preprint]. 2 May 2021. 10.1101/2020.11.02.20222778. [DOI]
- 25.Kundu R, Narean JS, Wang L, Fenn J, Pillay T, Fernandez ND, Conibear E, Koycheva A, Davies M, Tolosa-Wright M, Hakki S, Varro R, Dermott EM, Hammett S, Cutajar J, Thwaites RS, Parker E, Rosadas C, Clure MM, Tedder R, Taylor GP, Dunning J, Lalvani A, Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat. Commun. 13, 80 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katz JD, Bouley AJ, Jungquist RM, Douglas EA, O’Shea IL, Lathi ES, Humoral and T-cell responses to SARS-CoV-2 vaccination in multiple sclerosis patients treated with ocrelizumab. Mult. Scler. Relat. Disord. 57, 103382 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apostolidis SA, Kakara M, Painter MM, Goel RR, Mathew D, Lenzi K, Rezk A, Patterson KR, Espinoza DA, Kadri JC, Markowitz DM, Markowitz CE, Mexhitaj I, Jacobs D, Babb A, Betts MR, Prak ETL, Weiskopf D, Grifoni A, Lundgreen KA, Gouma S, Sette A, Bates P, Hensley SE, Greenplate AR, Wherry EJ, Li R, Bar-Or A, Cellular and humoral immune responses following SARS-CoV-2 mRNA vaccination in patients with multiple sclerosis on anti-CD20 therapy. Nat. Med. 27, 1990–2001 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bange EM, Han NA, Wileyto P, Kim JY, Gouma S, Robinson J, Greenplate AR, Hwee MA, Porterfield F, Owoyemi O, Naik K, Zheng C, Galantino M, Weisman AR, Ittner CAG, Kugler EM, Baxter AE, Oniyide O, Agyekum RS, Dunn TG, Jones TK, Giannini HM, Weirick ME, Allister CMM, Babady NE, Kumar A, Widman AJ, De Wolf S, Boutemine SR, Roberts C, Budzik KR, Tollett S, Wright C, Perloff T, Sun L, Mathew D, Giles JR, Oldridge CA, Wu JE, Alanio C, Adamski S, Garfall AL, Vella LA, Kerr SJ, Cohen JV, Oyer RA, Massa R, Maillard IP, Maxwell KN, Reilly JP, Maslak PG, Vonderheide RH, Wolchok JD, Hensley SE, Wherry EJ, Meyer NJ, De Michele AM, Vardhana SA, Mamtani R, Huang AC, CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 27, 1280–1289 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, Chng MHY, Lin M, Tan N, Linster M, Chia WN, Chen MI-C, Wang L-F, Ooi EE, Kalimuddin S, Tambyah PA, Low JG-H, Tan Y-J, Bertoletti A, SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584, 457–462 (2020). [DOI] [PubMed] [Google Scholar]
- 30.le Bert N, Clapham HE, Tan AT, Chia WN, Tham CYL, Lim JM, Kunasegaran K, Tan LWL, Dutertre C-A, Shankar N, Lim JME, Sun LJ, Zahari M, Tun ZM, Kumar V, Lim BL, Lim SH, Chia A, Tan Y-J, Tambyah PA, Kalimuddin S, Lye D, Low JGH, Wang L-F, Wan WY, Hsu LY, Bertoletti A, Tam CC, Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 218, e20202617 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, Dejnirattisai W, Rostron T, Supasa P, Liu C, López-Camacho C, Slon-Campos J, Zhao Y, Stuart DI, Paesen GC, Grimes JM, Antson AA, Bayfield OW, Hawkins DEDP, Ker D-S, Wang B, Turtle L, Subramaniam K, Thomson P, Zhang P, Dold C, Ratcliff J, Simmonds P, de Silva T, Sopp P, Wellington D, Rajapaksa U, Chen Y-L, Salio M, Napolitani G, Paes W, Borrow P, Kessler BM, Fry JW, Schwabe NF, Semple MG, Baillie JK, Moore SC, Openshaw PJM, Ansari MA, Dunachie S, Barnes E, Frater J, Kerr G, Goulder P, Lockett T, Levin R, Zhang Y, Jing R, Ho L-P ; Oxford Immunology Network Covid-19 Response T cell Consortium; ISARIC4C Investigators, Cornall RJ, Conlon CP, Klenerman P, Screaton GR, Mongkolsapaya J, Michael, Knight JC, Ogg G, Dong T, Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 21, 1336–1345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, Alanio C, Kuri-Cervantes L, Pampena MB, D’Andrea K, Manne S, Chen Z, Huang YJ, Reilly JP, Weisman AR, Ittner CAG, Kuthuru O, Dougherty J, Nzingha K, Han N, Kim J, Pattekar A, Goodwin EC, Anderson EM, Weirick ME, Gouma S, Arevalo CP, Bolton MJ, Chen F, Lacey SF, Ramage H, Cherry S, Hensley SE, Apostolidis SA, Huang AC, Vella LA; UPenn COVID Processing Unit, Betts MR, Meyer NJ, Wherry EJ, Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 369, eabc8511 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mallajosyula V, Ganjavi C, Chakraborty S, McSween AM, Pavlovitch-Bedzyk AJ, Wilhelmy J, Nau A, Manohar M, Nadeau KC, Davis MM, CD8+ T cells specific for conserved coronavirus epitopes correlate with milder disease in COVID-19 patients. Sci. Immunol. 6, eabg5669 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du J, Wei L, Li G, Hua M, Sun Y, Wang D, Han K, Yan Y, Song C, Song R, Zhang H, Han J, Liu J, Kong Y, Persistent high percentage of HLA-DR+CD38high CD8+ T cells associated with immune disorder and disease severity of COVID-19. Front. Immunol. 12, 735125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ameratunga R, Woon S-T, Jordan A, Longhurst H, Leung E, Steele R, Lehnert K, Snell R, Brooks AES, Perspective: Diagnostic laboratories should urgently develop T cell assays for SARS-CoV-2 infection. Expert Rev. Clin. Immunol. 17, 421–430 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Habel JR, Nguyen THO, van de Sandt CE, Juno JA, Chaurasia P, Wragg K, Koutsakos M, Hensen L, Jia X, Chua B, Zhang W, Tan H-X, Flanagan KL, Doolan DL, Torresi J, Chen W, Wakim LM, Cheng AC, Doherty PC, Petersen J, Rossjohn J, Wheatley AK, Kent SJ, Rowntree LC, Kedzierska K, Suboptimal SARS-CoV-2−specific CD8+ T cell response associated with the prominent HLA-A*02:01 phenotype. Proc. Natl. Acad. Sci. U.S.A. 117, 24384–24391 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalai SC, Dines JN, Snyder TM, Gittelman RM, Eerkes T, Vaney P, Howard S, Akers K, Skewis L, Monteforte A, Witte P, Wolf C, Nesse H, Herndon M, Qadeer J, Duffy S, Svejnoha E, Taromino C, Kaplan IM, Alsobrook J, Manley T, Baldo L, Clinical validation of a novel T-cell receptor sequencing assay for identification of recent or prior SARS-CoV-2 infection. medRxiv 2021.01.06.21249345 [Preprint]. 8 January 2021. 10.1101/2021.01.06.21249345. [DOI] [PMC free article] [PubMed]
- 38.Snyder TM, Gittelman RM, Klinger M, May DH, Osborne EJ, Taniguchi R, Jabran Zahid H, Kaplan IM, Dines JN, Noakes MT, Pandya R, Chen X, Elasady S, Svejnoha E, Ebert P, Pesesky MW, De Almeida P, O’Donnell H, De Gottardi Q, Keitany G, Lu J, Vong A, Elyanow R, Fields P, Greissl J, Baldo L, Semprini S, Cerchione C, Nicolini F, Mazza M, Delmonte OM, Dobbs K, Laguna-Goya R, Carreño-Tarragona G, Barrio S, Imberti L, Sottini A, Quiros-Roldan E, Rossi C, Biondi A, Bettini KR, D’Angio M, Bonfanti P, Tompkins MF, Alba C, Dalgard C, Sambri V, Martinelli G, Goldman JD, Heath JR, Su HC, Notarangelo LD, Paz-Artal E, Martinez-Lopez J, Carlson JM, Robins HS, Magnitude and dynamics of the T-cell response to SARS-CoV-2 infection at both individual and population levels. medRxiv 2020.07.31.20165647 [Preprint]. 17 September 2020. 10.1101/2020.07.31.20165647. [DOI]
- 39.Shoukat MS, Foers AD, Woodmansey S, Evans SC, Fowler A, Soilleux EJ, Use of machine learning to identify a T cell response to SARS-CoV-2. Cell Rep. Med. 2, 100192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.CDC, “Interferon-gamma release assays (IGRAs) – Blood tests for TB infection” (2011); www.cdc.gov/tb/publications/factsheets/testing/igra.htm.
- 41.Murugesan K, Jagannathan P, Pham TD, Pandey S, Bonilla HF, Jacobson K, Parsonnet J, Andrews JR, Weiskopf D, Sette A, Pinsky BA, Singh U, Banaei N, Interferon-γ release assay for accurate detection of SARS-CoV-2 T cell response. Clin. Infect. Dis. 73, e3130–e3132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.U.S. FDA, “Policy for Coronavirus Disease-2019 Tests During the Public Health Emergency (Revised)”; www.fda.gov/regulatoryinformation/search-fda-guidance-documents/policy-coronavirus-disease-2019-tests-during-publichealth-emergency-revised.
- 43.Levin EG, Lustig Y, Cohen C, Fluss R, Indenbaum V, Amit S, Doolman R, Asraf K, Mendelson E, Ziv A, Rubin C, Freedman L, Kreiss Y, Regev-Yochay G, Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N. Engl. J. Med. 385, e84 (2021). 44. “Emergency use authorization for vaccines to prevent COVID-19: Guidance for industry” (U.S. Food and Drug Administration/Center for Biologics Evaluation and Research, 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfizer press release; www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-provideupdate-ongoing-studies-covid-19.
- 45.Tada T, Zhou H, Samanovic MI, Dcosta BM, Cornelius A, Mulligan MJ, Landau NR, Comparison of neutralizing antibody titers elicited by mRNA and adenoviral vector vaccine against SARS-CoV-2 variants. bioRxiv 2021.07.19.452771 [Preprint]. 19 July 2021. 10.1101/2021.07.19.452771. [DOI]
- 46.Barouch DH, Stephenson KE, Sadoff J, Yu J, Chang A, Gebre M, Mahan KM, Liu J, Chandrashekar A, Patel S, Gars ML, de Groot AM, Heerwegh D, Struyf F, Douoguih M, van Hoof J, Schuitemaker H, Durable humoral and cellular immune responses 8 months after Ad26.COV2.S vaccination. N. Engl. J. Med. 385, 951–953 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ford CT, Machado DJ, Janies DA, Predictions of the SARS-CoV-2 Omicron variant (B.1.1.529) spike protein receptor-binding domain structure and neutralizing antibody interactions. bioRxiv 10.3389/fviro.2022.830202 [Preprint]. 7 February 2022. 10.3389/fviro.2022.830202. [DOI]
- 48.Zeng C, Evans JP, King T, SARS-CoV-2 spreads through cell-to-cell transmission. Proc. Natl. Acad. Sci. U.S.A. 119, e2111400119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]