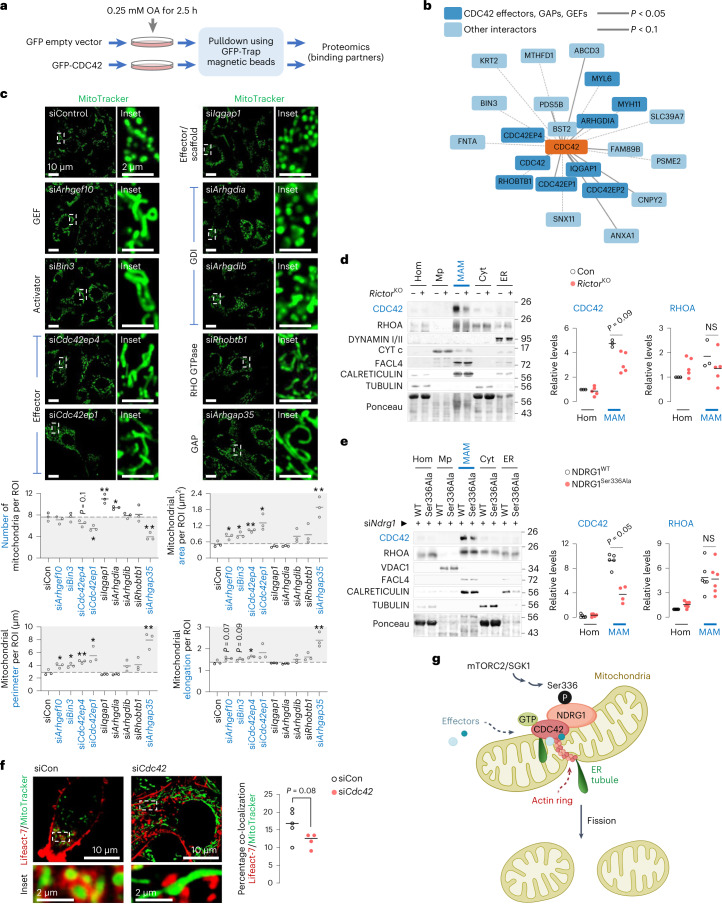
Fig. 7. CDC42 regulators and effectors modulate fission.
a, Experimental plan to pull down GFP-tagged CDC42 to identify interactors that regulate mitochondrial fission. b, Cartoon representing significantly (P < 0.05) enriched interacting partners of CDC42 (in bold) identified via proteomics, some of which belong to the RHO family of GTPases (n = 4 independent experiments). c, Representative images of NIH3T3 cells knocked-down for indicated CDC42-binding partners and cultured in serum-free medium for 30 min in presence of MitoTracker green. Magnified insets are shown. Quantifications for mitochondrial number and mitochondrial size/shape descriptors are shown (siCon, siArhgef10, siIqgap, siArhgdib, siRhobtb1 and siArhgap35 39 cells, siBin3 and siCdc42ep4 40 cells, siCdc42ep1 43 cells and siArhgdia 37 cells from n = 3 independent experiments). Grey areas indicate mitochondria fission-deficient models. d,e, Immunoblots and quantification for indicated proteins in indicated fractions from livers of 14–16 h-fasted 5–6-month-old Con (n = 3) and RictorKO (n = 5) mice (d), and 3–4-month-old mice expressing NDRG1WT or NDRG1Ser336Ala plasmids after silencing endogenous Ndrg1 with siRNAs (e). N values for number of mice per fraction are indicated in parentheses. CDC42: all fractions from NDRG1WT and NDRG1Ser336Ala mice are n = 5 except for NDRG1Ser336Ala MAMs where n = 4; RHOA: n = 6 mice for all groups. Ponceau is loading control. f, Representative confocal images of siCon or siCdc42 NIH3T3 cells expressing mCherry–Lifeact-7 and cultured in serum-free medium for 30 min in presence of MitoTracker green. Magnified insets are shown. Quantification for percentage co-localization of mCherry–Lifeact-7 with mitochondria is shown (siCon 33 cells and siCdc42 30 cells from n = 5 (siCon) or n = 4 (siCdc42) independent experiments). g, Reactivation of mTORC2 during fasting phosphorylates NDRG1 at Ser336, which engages with mitochondria and recruits CDC42 to mitochondria–ER contact sites wherein CDC42 and its effector proteins orchestrate fission. Individual replicates and means are shown. *P < 0.05 and **P < 0.01, two-tailed unpaired Student’s t-test. NS, not significant. Please refer to Supplementary Table 10 statistical summary, and Supplementary Table 9. Source numerical data are available in Source Data Extended Data Table 1, and unprocessed blots are available in the Source Data for this figure.