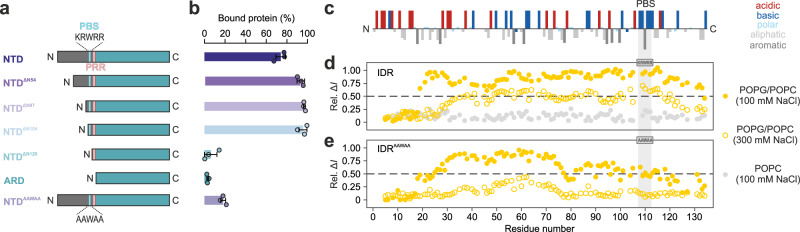
Fig. 7. Extensive lipid binding in the IDR is negatively affected by the distal N-terminus.
a Topology of N-terminal deletion mutants used for liposome sedimentation assay. b Protein distribution between pellet (“bound protein”) or supernatant fraction after centrifugation, quantified via densitometry of SDS-PAGE protein bands using imageJ51. Data are presented as the mean value ± SEM from n = 3 individual experiments. c Distribution of charged and hydrophobic residues in the TRPV4-IDR shows a gradient of a consecutively more basic and hydrophobic protein from N- to C-terminus. Plotted with the PepCalc tool (https://pepcalc.com/). d, e NMR signal intensity differences for 15N-labeled IDR variants (100 µM) in the absence and presence of POPC (light gray circles) or POPC-POPG containing liposomes at low (filled yellow circles) or high salt concentration (open yellow circles). Higher values are indicative of lipid binding. Source data are provided as a Source Data file.