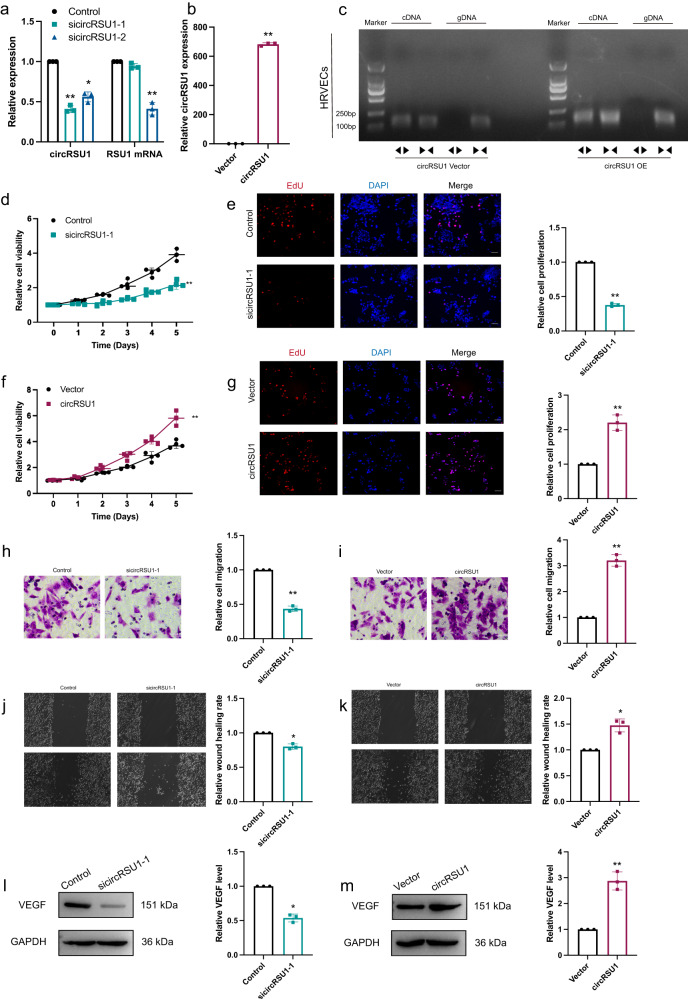
Fig. 2. circRSU1 promoted DR progression.
a, b The efficiency of circRSU1 interference was determined by qRT-PCR. c PCR analysis revealed the presence of circRSU1 overexpression (OE) through backsplicing. A negative control with circRSU1 vector was utilized. d–g. MTT assay and EdU staining were performed to detect the proliferation of HRVECs (scale bar = 100 μm). h, i Transwell invasion assay was used to detect the invasion ability of HRVECs (scale bar = 20 μm). j, k. Wound-healing migration assay was performed to detect the migration ability of HRVECs (scale bar = 100 μm). l, m Western blot was used to analyze VEGF expression. GAPDH was used as a negative control. Unpaired student’s t-test, Mann–Whitney U test and one-way ANOVA test were used for the statistical analyses. *p < 0.05,**p < 0.01.