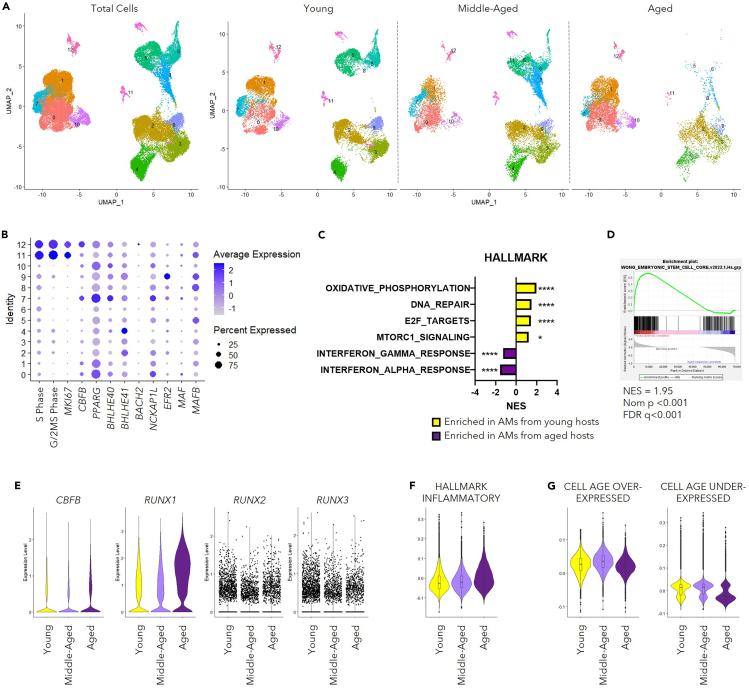
Figure 5.
AMs from the elderly possess senescence-like phenotypes
Data of human AMs were extracted from combined scRNA-seq profiling pneumocytes of “Healthy” donors (GSE135893,51GSE122960,52GSE128033,53GSE13683154). Samples were divided into “Young” (<40-years old), “Middle-Aged” (40-60- years old) and “Aged” (≥60-years old) groups. In total, 79 samples with 35 “Young”, 27 “Middle-Aged”, and 17 ″Aged” were included.
(A) UMAP displaying the clustering of AMs as total cells (left) and in separate groups (right).
(B) Dot plot revealing the score for S-Phase or G/2M-Phase related genes, transcription factors related to proliferation and identity of AMs, and transcription factors associated with the suppression of the ESC-like feature of AMs.
(C) Bar graph displaying selected pathways identified with GSEA using gene sets from HALLMARK database comparing individuals from “Young” and “Aged” groups.
(D) GSEA result analysis with a list of genes associated with ESC-like features (MSigDB M7079).
(E) Violin Plots displaying mRNA level of CBFB and RUNX family members (RUNX1, RUNX2, and RUNX3).
(F) Violin Plot displaying module scores calculated with a gene list featuring inflammatory response (MSigDB M5932).
(G) Violin plots revealing module scores calculated with gene lists associated with cellular senescence.40
Data in (B) were displayed in dots with diameter representing the percent of cells in the cluster (as shown in A) that expressed the gene (or list of genes) indicated, with the depth of color indicating the average expression level. Data in (C and D) were analyzed with GSEA, shown as ∗, p < 0.05; ∗∗∗∗, p < 0.0001. The boxplots within the Violin Plots in (F) and (G) displayed the 25, 50 and 75 percentiles of the module scores calculated with genes lists indicated. See also Figure S5.