Figure 1.
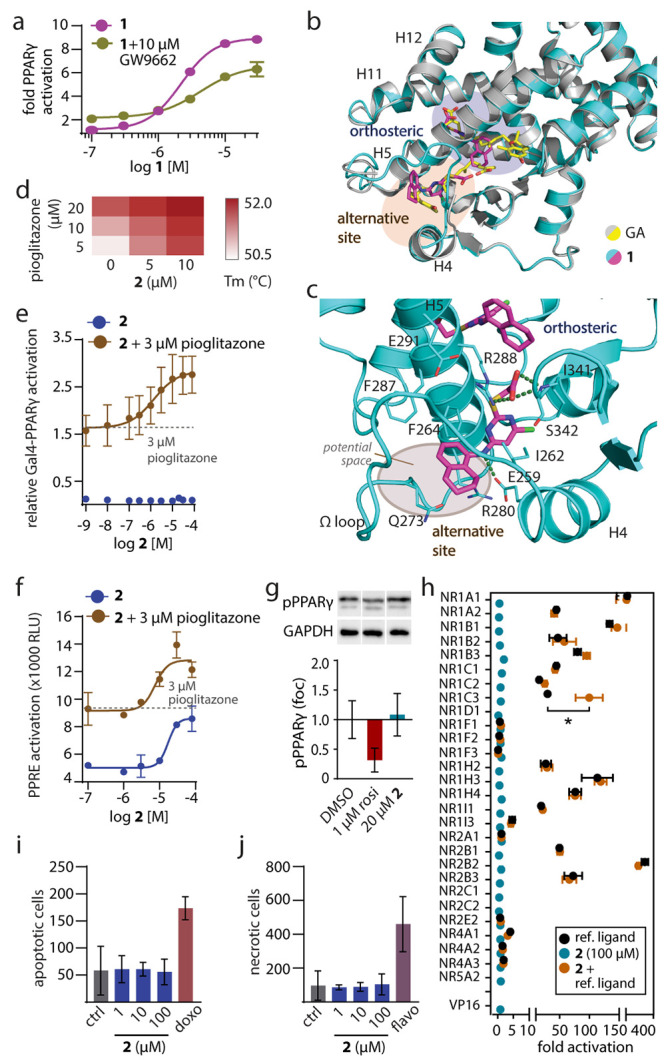
Characterization of 1 mimicking the dual PPARγ binding of GA and of the selective alternative site PPARγ ligand 2. (a) Dose–response curves of 1 in a Gal4-PPARγ reporter gene assay in the absence and presence of the irreversible orthosteric antagonist GW9662 (10 μM). Data are the mean ± S.E.M., n = 3. (b) The X-ray structure of the PPARγ LBD in complex with 1 (pdb ID: 8aty) revealed two molecules binding to the LBD and highly aligned with the PPARγ-GA complex (pdb ID: 7awd).12 Electron density map for 1 in Figure S2. (c) Second site binding of 1 and GA revealed an interaction with the side chain of Arg288 and potential space for an extension of 1 to achieve selective binding to this site. (d) Thermal stability of the PPARγ LBD in the presence of different concentrations of 2 and pioglitazone. The heat map shows the mean Tm, n = 3. (e, f) Effects of 2 on Gal4-PPARγ (e) and PPRE (f) activity in the absence and presence of pioglitazone. Data are the mean ± S.E.M., n ≥ 3. (g) Effects of 2 on PPARγ Ser273 phosphorylation. Data are the mean ± S.E.M., n = 3–6. Rosiglitazone as positive control. Western blots in Supporting Information. (h) Selectivity profiling of 2 on nuclear receptors. Data are the mean ± S.E.M., n ≥ 3. * p < 0.05 (t-test 2 vs 2 + ref. ligand). (i, j) Effects of 2 on apoptosis (i) and necrosis (j) in COS7 cells after 24 h. Doxorubicin (doxo, 100 μM) and flavopiridol (flavo, 100 μM) as positive controls. Data are the mean ± S.E.M., n = 3.
