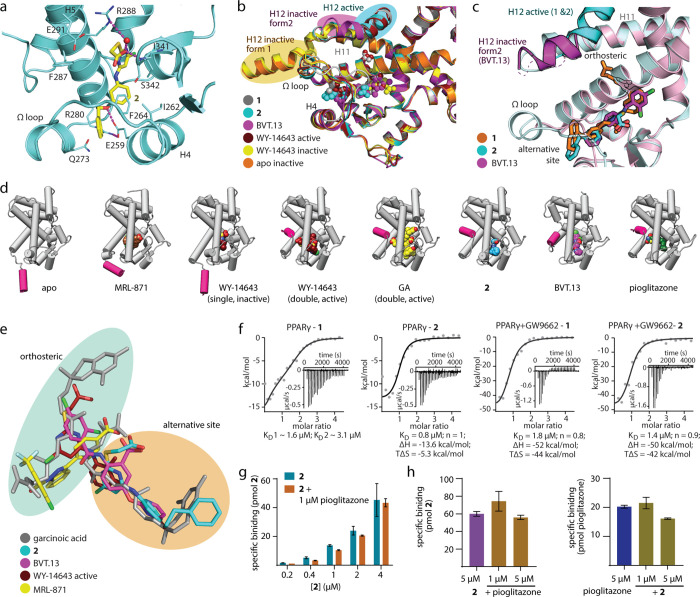
Figure 2.
Binding of 2 to PPARγ (pdb ID: 8atz). (a) The co-crystal structure of the PPARγ LBD in complex with 2 (pdb ID: 8atz) revealed selective binding of the ligand to the alternative site and a stabilized active conformation of helix 12. Binding of 2 to the PPARγ LBD was mediated by H-bonds to Arg288 and Ser342. Electron density map for 2 in Figure S2. (b) PPARγ ligands 1, 2, BVT.13 (pdb ID: 2q6s11), and WY14643 (pdb ID: 8cph, 8cpi) induce different conformations of helix 12. (c) Compared to BVT.13, binding of 2 is shifted outward from the orthosteric region. (d) Comparison of PPARγ LBD structures in complex with various alternative sites and double binding ligands reveals differences in ligand binding sites and conformations. Apo (pdb ID: 8cpj) and pioglitazone-complexed (pdb ID: 5y2o27) PPARγ structures are shown as representatives of inactive and active forms, respectively. (e) Superposition of the bound ligands GA, 2, BVT.13, MRL-871, and WY14643 demonstrates different binding modes within the orthosteric and alternative binding regions. (f) Isothermal titration calorimetry (ITC) for the binding of 1 and 2 to the ligand-free and the GW9662-bound PPARγ LBD. The fitting of the heat of binding is shown with the isotherms as insets. (g, h) LC–MS-based binding experiments demonstrated dose-dependent specific binding of 2 (0.2–4 μM) to the PPARγ LBD (1 μM) in the absence and presence of 1 μM pioglitazone (g) and specific binding of 2 and pioglitazone (5 μM each) in the absence or presence of the respective other ligand (h); data are the mean ± S.E.M., n = 3.