Abstract
Firefly luciferin methyl ester is hydrolyzed by monoacylglycerol lipase MAGL, amidase FAAH, poorly-characterized hydrolase ABHD11, and hydrolases known for S-depalmitoylation (LYPLA1/2), not just esterase CES1. This enables activity-based bioluminescent assays for serine hydrolases and suggests that the ‘esterase activity’ responsible for hydrolyzing ester prodrugs is more diverse than previously supposed.
Fireflies are beetles that emit a green glow when the enzyme firefly luciferase adenylates and oxidizes the small molecule d-luciferin. The chemistry of bioluminescence requires a free carboxylic acid, and beetle luciferases evolved from enzymes that adenylate fatty acids (Figure 1).1–3 Embracing d-luciferin and analogues as fatty-acid mimics previously led us to design luciferin amides as probes that specifically report on the enzymatic activity of fatty acid amide hydrolase (FAAH),4 a membrane-bound enzyme that hydrolyzes fatty acid amide neurotransmitters such as anandamide and oleamide, and has been a popular target for pharmaceutical development.5,6 Hydrolysis of luciferin amides in the presence of firefly luciferase results in bioluminescence, enabling detection of FAAH activity in live cells and in vivo.4,7–9 Building on this concept, we hypothesized that luciferin esters could similarly be substrates for lipases that regulate signaling lipids, in particular monoacylglycerol lipase (MAGL, aka MGLL) and the alpha/beta hydrolase domain (ABHD) enzymes ABHD6 and ABHD12, as these enzymes all hydrolyze the endocannabinoid 2-arachidonoyl glycerol (2-AG) to arachidonic acid (Figure 1).10
Figure 1.
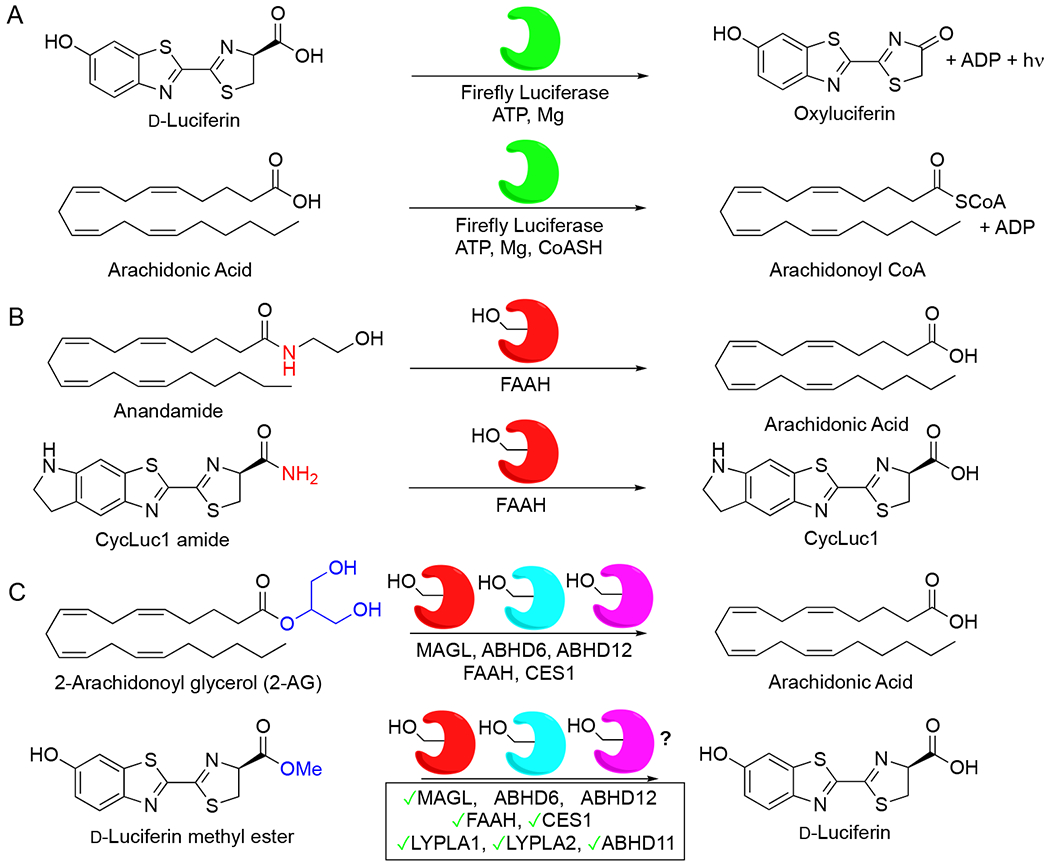
Building bioluminescent sensors for serine hydrolases. A) Firefly luciferase oxidizes d-luciferin to emit light, but also has fatty acyl-CoA synthetase activity; B) In previous work, we embraced d-luciferin and luciferin analogues as fatty acid mimics, finding that luciferin amides can be hydrolyzed by FAAH, forming the basis for bioluminescent detection of FAAH activity; C) Here, we propose that d-luciferin methyl ester can similarly serve as a sensor of monoacylglycerol lipase activity.
Esters of d-luciferin have long been known to be cell-permeable and hydrolyzed by “esterase activity”,11,12 but the specific enzyme(s) responsible for ester hydrolysis have not been well described. It has been reported that d-luciferin methyl ester is specifically hydrolyzed by human carboxyesterase 1 (CES1).13 However, we have found that ethyl esters of d-luciferin and the luciferin analogue CycLuc1 can be hydrolyzed by the amidase FAAH,4 and many mammalian cells do not appreciably express CES1 or FAAH, yet still hydrolyze esters.14,15 Here we show that d-luciferin methyl ester is a substrate for a number of serine hydrolases, including MAGL, lysophosphoplipase 1 (LYPLA1), lysophospholipase 2 (LYPLA2), and ABHD11.16 Activity-based bioluminescence assays17 for these hydrolases are thus enabled, complementing activity-based protein profiling (ABPP) methods,18 and adding the ability to study serine hydrolases in live cells. More broadly, these results also suggest that the scope of enzymes capable of hydrolyzing ester prodrugs extends beyond the canonical carboxyesterase family,19,20 with implications for their selective unmasking in different cell types.
Human PC3 prostate and U87 glioma cell lines both express MAGL and ABHD6, but not CES1 or FAAH.14,15,21,22 We therefore transfected PC3 and U87 cells with firefly luciferase, then treated them with 10 μM d-luciferin methyl ester. A strong bioluminescent signal was observed from both cell lines, exceeding that of 10 μM d-luciferin itself (Figure S1),11 indicating that hydrolysis of the ester to the free carboxylate had occurred (Figure 2). We observed a significant and dose- dependent reduction in bioluminescence from cells pre-incubated with the MAGL inhibitors JZL184 and KML29,23 suggesting that MAGL contributes to this hydrolytic activity. Treatment of cells with the nonspecific serine protease inhibitor MAFP resulted in an even greater reduction in bioluminescence than KML29, suggesting that, in addition to MAGL, other cellular hydrolases could also cleave luciferin esters. However, the ABHD6 inhibitor24 KT182 did not inhibit bioluminescence, indicating that ABHD6 was not responsible, and the CES1 inhibitor25 WWL113 similarly had no effect, confirming the lack of CES1 involvement in these cells.
Figure 2.
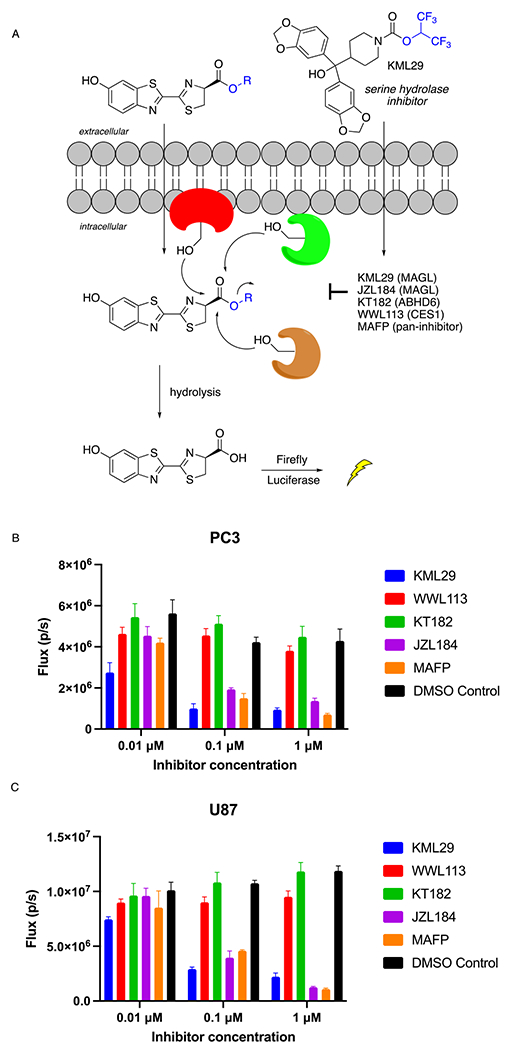
Bioluminescence from luciferase-transfected cells after d-luciferin methyl ester treatment. A) Hydrolysis of the methyl ester generates d-luciferin, resulting in bioluminescence. Inhibition of the enzyme(s) responsible for hydrolysis inhibits the bioluminescent signal; B,C) Luciferase-expressing PC3 prostate cells (B) and U87 glioma cells (C) were treated with the indicated serine hydrolase inhibitors, then imaged with d-luciferin methyl ester (10 μM).
To better tease apart which enzymes are capable of hydrolyzing d-luciferin methyl ester, we expressed a panel of candidate hydrolases in HEK293 cells. We focused on human serine hydrolases that have demonstrated activity toward fatty acyl glycerols (MAGL, ABHD6, ABHD12, FAAH, CES1),10 but also included several serine hydrolases that are less well characterized. LYPLA1 and LYPLA2,26 also known as acyl-protein thioesterase (APT) 1 and 2, are perhaps best known for hydrolyzing palmitate thioesters from proteins.27,28 The reported ability of LYPLA2 to hydrolyze prostaglandin glycerol esters29 suggested to us that it may also be capable of hydrolyzing luciferin esters. In addition, we included ABHD11, a poorly-characterized serine hydrolase that is presumed to act on lipids, but whose endogenous substrate(s) have not been firmly established.30–34 As controls, we expressed the corresponding mutant enzymes in which the key catalytic serine residue was mutated to alanine. When co-expressed with firefly luciferase, a number of these hydrolytic enzymes demonstrated an ability to enhance bioluminescence from d-luciferin methyl ester compared to their control (Figure 3, Figure S2). Active hydrolase expression had no effect on bioluminescence from d-luciferin (Figure S3). These results confirmed that, in addition to CES1 and FAAH,4,13 the hydrolytic activity of MAGL can cleave d-luciferin methyl ester. Moreover, LYPLA1, LYPLA2, and, unexpectedly, ABHD11, were also found to greatly increase the bioluminescent signal. We expect that this list is not exhaustive. On the other hand, the monoacylglycerol lipases ABHD6 and ABHD12 surprisingly showed no activity toward d-luciferin methyl ester (Figure 3) despite their known activity toward 2-AG (Figure 1).10 It is possible that active ABHD6 and ABHD12 are poorly expressed in our assay, but the previous successful expression of these enzymes suggests that this is not the case.35,36 Moreover, the ABHD6 results are consistent with the inability of KT182 to block bioluminescence from d-luciferin methyl ester in PC3 and U87 cells (Figure 2).
Figure 3.
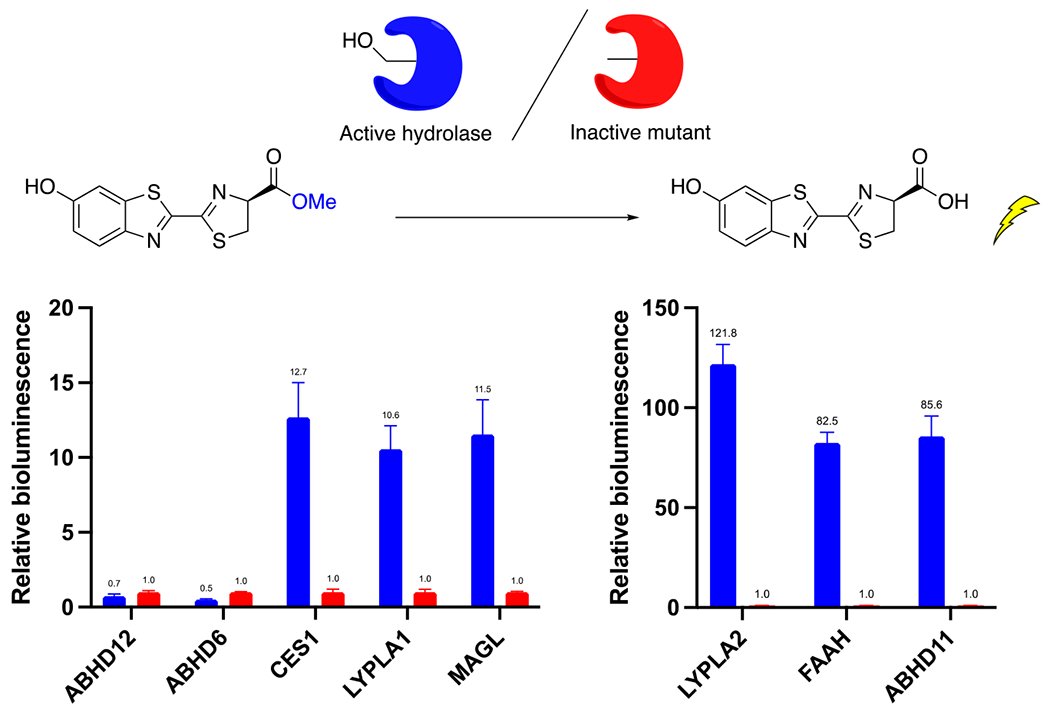
Effect of serine hydrolase expression on bioluminescence with d-luciferin methyl ester in HEK293 cells. Represented as the fold-increase in bioluminescence for the active serine hydrolase compared to its control inactive serine to alanine mutant.
We next asked whether d-luciferin methyl ester could function as an activity-based bioluminescence probe for detecting the inhibition of serine hydrolases overexpressed in HEK293 cells without interference from endogenous hydrolase activity. ML226 has been reported to be a specific inhibitor of ABHD11.37 We found that ML226 potently inhibits the bioluminescent signal from d-luciferin methyl ester in cells expressing ABHD11 in a dose-dependent manner, bringing the bioluminescence down to the level of the inactive S141A ABHD11 control, whereas the MAGL inhibitors KML29 and JZL184 have no effect (Figure 4). This suggests that d-luciferin methyl ester can be used to identify ABHD11 inhibitors using this live cell assay, and that this general strategy has potential for the identification and characterization of inhibitors of other serine hydrolases.
Figure 4.

Bioluminescence emission from HEK293 cells expressing ABHD11 and firefly luciferase treated with d-luciferin methyl ester and the indicated serine hydrolase inhibitors, compared to the inactive ABHD11-S141A mutant control.
Our results indicate that d-luciferin methyl ester is not just a substrate for CES1 and suggests that luciferin esters in general hold promise as probes for the detection of numerous serine hydrolases, particularly those with lipid substrates. d-Luciferin methyl ester is not as specific for lipases as d-luciferin and CycLuc1 amides are for FAAH, and a challenge for the future translation of luciferin esters to in vivo imaging applications is the promiscuous activity of CES1. Nonetheless, d-luciferin methyl ester can presently be used to selectively detect the endogenous activity of MAGL in live cells. Furthermore, by expressing active and inactive forms of hydrolytic enzymes, the activity from other lipases such as ABHD11 can be detected, enabling the identification and characterization of inhibitors in situ. Many serine hydrolases are membrane-bound and remain poorly characterized,16,38 and even those that have been the subject of study continue to reveal new surprises.39 New tools to identify and validate the selectivity and specificity40 of inhibitors in live cells are thus valuable. Given the ever-expanding scope of luminogenic luciferin analogues,8,41–44 we anticipate that modified luciferin structures as well as the incorporation of different esters can be utilized in combination to further refine the specificity and expand the scope of activity-based bioluminescence probes for enzymes that process lipid substrates, potentially including ABHD6 and ABHD12, among others. Finally, our results indicate that the hydrolytic activity typically ascribed to “esterases” can also arise from several serine hydrolases that are involved in specialized aspects of lipid metabolism. This includes, but is not limited to, FAAH, MAGL, LYPLA1, LYPLA2, and ABHD11. Notably, the use of FAAH for amide prodrug activation has recently been reported to extend beyond luciferin analogues.45 The activity of serine hydrolases like MAGL toward ester prodrugs may similarly have implications for their subsequent unmasking in different cell types.
Supplementary Material
Acknowledgements
This work was supported by the U.S. National Institutes of Health (DA039961 and EB013270). I.M. performed synthesis and HEK cell assays; K.L.L. performed PC3 and U87 cell assays; S.C.M. conceived the project, directed research, and wrote the paper with input from I.M.
References
- (1).Oba Y; Sato M; Ojika M; Inouye S Enzymatic and Genetic Characterization of Firefly Luciferase and Drosophila CG6178 as a Fatty Acyl-CoA Synthetase. Biosci Biotechnol Biochem 2005, 69 (4), 819–828. [DOI] [PubMed] [Google Scholar]
- (2).Fallon TR; Lower SE; Chang C-H; Bessho-Uehara M; Martin GJ; Bewick AJ; Behringer M; Debat HJ; Wong I; Day JC; Suvorov A; Silva CJ; Stanger-Hall KF; Hall DW; Schmitz RJ; Nelson DR; Lewis SM; Shigenobu S; Bybee SM; Larracuente AM; Oba Y; Weng J-K Firefly Genomes Illuminate Parallel Origins of Bioluminescence in Beetles. eLife 2018, 7, e36495. 10.7554/eLife.36495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Adams ST; Miller SC Enzymatic Promiscuity and the Evolution of Bioluminescence. The FEBS Journal 2020, 287 (7), 1369–1380. 10.1111/febs.15176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mofford DM; Adams ST; Reddy GSKK; Reddy GR; Miller SC Luciferin Amides Enable in Vivo Bioluminescence Detection of Endogenous Fatty Acid Amide Hydrolase Activity. J. Am. Chem. Soc 2015, 137 (27), 8684–8687. 10.1021/jacs.5b04357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Cravatt BF; Giang DK; Mayfield SP; Boger DL; Lerner RA; Gilula NB Molecular Characterization of an Enzyme That Degrades Neuromodulatory Fatty-Acid Amides. Nature 1996, 384 (6604), 83–87. 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- (6).Ahn K; Johnson DS; Mileni M; Beidler D; Long JZ; McKinney MK; Weerapana E; Sadagopan N; Liimatta M; Smith SE; Lazerwith S; Stiff C; Kamtekar S; Bhattacharya K; Zhang Y; Swaney S; Van Becelaere K; Stevens RC; Cravatt BF Discovery and Characterization of a Highly Selective FAAH Inhibitor That Reduces Inflammatory Pain. Chem. Biol 2009, 16 (4), 411–420. 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Adams ST; Mofford DM; Reddy GSKK; Miller SC Firefly Luciferase Mutants Allow Substrate-Selective Bioluminescence Imaging in the Mouse Brain. Angew. Chem. Int. Ed 2016, 55, 4943–4946. 10.1002/anie.201511350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Miller SC; Mofford DM; Adams ST Lessons Learned from Luminous Luciferins and Latent Luciferases. ACS Chem. Biol 2018, 13 (7), 1734–1740. 10.1021/acschembio.7b00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ji X; Adams ST; Miller SC Chapter Eight - Bioluminescence Imaging in Mice with Synthetic Luciferin Analogues. In Methods in Enzymology; Chenoweth DM, Ed.; Chemical Tools for Imaging, Manipulating, and Tracking Biological Systems: Diverse Methods Based on Optical Imaging and Fluorescence; Academic Press, 2020; Vol. 640, pp 165–183. 10.1016/bs.mie.2020.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Blankman JL; Simon GM; Cravatt BF A Comprehensive Profile of Brain Enzymes That Hydrolyze the Endocannabinoid 2-Arachidonoylglycerol. Chem. Biol 2007, 14 (12), 1347–1356. 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Craig FF; Simmonds AC; Watmore D; McCapra F; White MR Membrane-Permeable Luciferin Esters for Assay of Firefly Luciferase in Live Intact Cells. Biochem. J 1991, 276 ( Pt 3), 637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Miska W; Geiger R Synthesis and Characterization of Luciferin Derivatives for Use in Bioluminescence Enhanced Enzyme Immunoassays. New Ultrasensitive Detection Systems for Enzyme Immunoassays, I. J. Clin. Chem. Clin. Biochem 1987, 25 (1), 23–30. [DOI] [PubMed] [Google Scholar]
- (13).Wang D-D; Jin Q; Zou L-W; Hou J; Lv X; Lei W; Cheng H-L; Ge G-B; Yang L A Bioluminescent Sensor for Highly Selective and Sensitive Detection of Human Carboxylesterase 1 in Complex Biological Samples. Chem. Commun 2016, 52 (15), 3183–3186. 10.1039/C5CC09874B. [DOI] [PubMed] [Google Scholar]
- (14).Uhlén M; Fagerberg L; Hallström BM; Lindskog C; Oksvold P; Mardinoglu A; Sivertsson Å; Kampf C; Sjöstedt E; Asplund A; Olsson I; Edlund K; Lundberg E; Navani S; Szigyarto CA-K; Odeberg J; Djureinovic D; Takanen JO; Hober S; Alm T; Edqvist P-H; Berling H; Tegel H; Mulder J; Rockberg J; Nilsson P; Schwenk JM; Hamsten M; von Feilitzen K; Forsberg M; Persson L; Johansson F; Zwahlen M; von Heijne G; Nielsen J; Pontén F Tissue-Based Map of the Human Proteome. Science 2015, 347 (6220), 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- (15).Cell atlas - CES1 - The Human Protein Atlas. https://www.proteinatlas.org/ENSG00000198848-CES1/cell (accessed 2020–03-24).
- (16).Long JZ; Cravatt BF The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease. Chem. Rev 2011, 111 (10), 6022–6063. 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Su TA; Bruemmer KJ; Chang CJ Caged Luciferins for Bioluminescent Activity-Based Sensing. Current Opinion in Biotechnology 2019, 60, 198–204. 10.1016/j.copbio.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Liu Y; Patricelli MP; Cravatt BF Activity-Based Protein Profiling: The Serine Hydrolases. PNAS 1999, 96 (26), 14694–14699. 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Meyer MR; Schütz A; Maurer HH Contribution of Human Esterases to the Metabolism of Selected Drugs of Abuse. Toxicology Letters 2015, 232 (1), 159–166. 10.1016/j.toxlet.2014.10.026. [DOI] [PubMed] [Google Scholar]
- (20).Wang D; Zou L; Jin Q; Hou J; Ge G; Yang L Human Carboxylesterases: A Comprehensive Review. Acta Pharmaceutica Sinica B 2018, 8 (5), 699–712. 10.1016/j.apsb.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Cell atlas - MGLL - The Human Protein Atlas. https://www.proteinatlas.org/ENSG00000074416-MGLL/cell (accessed 2020–03-19).
- (22).Nomura DK; Lombardi DP; Chang JW; Niessen S; Ward AM; Long JZ; Hoover HH; Cravatt BF Monoacylglycerol Lipase Exerts Dual Control over Endocannabinoid and Fatty Acid Pathways to Support Prostate Cancer. Chemistry & Biology 2011, 18 (7), 846–856. 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Chang JW; Niphakis MJ; Lum KM; Cognetta AB; Wang C; Matthews ML; Niessen S; Buczynski MW; Parsons LH; Cravatt BF Highly Selective Inhibitors of Monoacylglycerol Lipase Bearing a Reactive Group That Is Bioisosteric with Endocannabinoid Substrates. Chemistry & Biology 2012, 19 (5), 579–588. 10.1016/j.chembiol.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hsu K-L; Tsuboi K; Chang JW; Whitby LR; Speers AE; Pugh H; Cravatt BF Discovery and Optimization of Piperidyl-1,2,3-Triazole Ureas as Potent, Selective, and in Vivo-Active Inhibitors of α/β-Hydrolase Domain Containing 6 (ABHD6). J. Med. Chem 2013, 56 (21), 8270–8279. 10.1021/jm400899c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dominguez E; Galmozzi A; Chang JW; Hsu K-L; Pawlak J; Li W; Godio C; Thomas J; Partida D; Niessen S; O’Brien PE; Russell AP; Watt MJ; Nomura DK; Cravatt BF; Saez E Integrated Phenotypic and Activity-Based Profiling Links Ces3 to Obesity and Diabetes. Nat. Chem. Biol 2014, 10 (2), 113–121. 10.1038/nchembio.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Wepy JA; Galligan JJ; Kingsley PJ; Xu S; Goodman MC; Tallman KA; Rouzer CA; Marnett LJ Lysophospholipases Cooperate to Mediate Lipid Homeostasis and Lysophospholipid Signaling. J. Lipid Res 2019, 60 (2), 360–374. 10.1194/jlr.M087890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Won SJ; Davda D; Labby KJ; Hwang SY; Pricer R; Majmudar JD; Armacost KA; Rodriguez LA; Rodriguez CL; Chong FS; Torossian KA; Palakurthi J; Hur ES; Meagher JL; Brooks CL; Stuckey JA; Martin BR Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2). ACS Chem. Biol 2016, 11 (12), 3374–3382. 10.1021/acschembio.6b00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Amara N; Foe IT; Onguka O; Garland M; Bogyo M Synthetic Fluorogenic Peptides Reveal Dynamic Substrate Specificity of Depalmitoylases. Cell Chemical Biology 2019, 26 (1), 35–47.e7. 10.1016/j.chembiol.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Manna JD; Wepy JA; Hsu K-L; Chang JW; Cravatt BF; Marnett LJ Identification of the Major Prostaglandin Glycerol Ester Hydrolase in Human Cancer Cells. J. Biol. Chem 2014, 289 (49), 33741–33753. 10.1074/jbc.M114.582353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Vijayakumar A; Vijayaraj P; Vijayakumar AK; Rajasekharan R The Arabidopsis ABHD11 Mutant Accumulates Polar Lipids in Leaves as a Consequence of Absent Acylhydrolase Activity. Plant Physiology 2016, 170 (1), 180–193. 10.1104/pp.15.01615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Arya M; Srinivasan M; Rajasekharan R Human Alpha Beta Hydrolase Domain Containing Protein 11 and Its Yeast Homolog Are Lipid Hydrolases. Biochemical and Biophysical Research Communications 2017, 487 (4), 875–880. 10.1016/j.bbrc.2017.04.145. [DOI] [PubMed] [Google Scholar]
- (32).Adibekian A; Martin BR; Wang C; Hsu K-L; Bachovchin DA; Niessen S; Hoover H; Cravatt BF Click-Generated Triazole Ureas as Ultrapotent in Vivo–Active Serine Hydrolase Inhibitors. Nature Chemical Biology 2011, 7 (7), 469–478. 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Bailey PSJ; Ortmann BM; Martinelli AW; Houghton JW; Costa ASH; Burr SP; Antrobus R; Frezza C; Nathan JA ABHD11 Maintains 2-Oxoglutarate Metabolism by Preserving Functional Lipoylation of the 2-Oxoglutarate Dehydrogenase Complex. Nature Communications 2020, 11 (1), 4046. 10.1038/s41467-020-17862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Escoubet J; Kenigsberg M; Derock M; Yaligara V; Bock M-D; Roche S; Massey F; Foucauld H. de; Bettembourg C; Olivier A; Berthemy A; Capdevielle J; Legoux R; Perret E; Buzy A; Chardenot P; Destelle V; Leroy A; Cahours C; Teixeira S; Juvet P; Gauthier P; Leguet M; Rocheteau-Beaujouan L; Chatoux M-A; Deshayes W; Clement M; Kabiri M; Orsini C; Mikol V; Didier M; Guillemot J-C ABHD11, a New Diacylglycerol Lipase Involved in Weight Gain Regulation. PLOS ONE 2020, 15 (6), e0234780. 10.1371/journal.pone.0234780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Navia-Paldanius D; Savinainen JR; Laitinen JT Biochemical and Pharmacological Characterization of Human α/β-Hydrolase Domain Containing 6 (ABHD6) and 12 (ABHD12). J. Lipid Res 2012, 53 (11), 2413–2424. 10.1194/jlr.M030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Yokoi N; Fukata Y; Sekiya A; Murakami T; Kobayashi K; Fukata M Identification of PSD-95 Depalmitoylating Enzymes. J. Neurosci 2016, 36 (24), 6431–6444. 10.1523/JNEUROSCI.0419-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Adibekian A; Hsu K-L; Speers AE; Brown SJ; Spicer T; Fernandez-Vega V; Ferguson J; Cravatt BF; Hodder P; Rosen H Optimization and Characterization of a Triazole Urea Inhibitor for Alpha/Beta Hydrolase Domain-Containing Protein 11 (ABHD11): Anti-Probe for LYPLA1/LYPLA2 Dual Inhibitor ML211. In Probe Reports from the NIH Molecular Libraries Program; National Center for Biotechnology Information (US): Bethesda (MD), 2010. [PubMed] [Google Scholar]
- (38).Lord CC; Thomas G; Brown JM Mammalian Alpha Beta Hydrolase Domain (ABHD) Proteins: Lipid Metabolizing Enzymes at the Interface of Cell Signaling and Energy Metabolism. Biochim Biophys Acta 2013, 1831 (4), 792–802. 10.1016/j.bbalip.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Cao Y; Qiu T; Kathayat RS; Azizi S-A; Thorne AK; Ahn D; Fukata Y; Fukata M; Rice PA; Dickinson BC ABHD10 Is an S -Depalmitoylase Affecting Redox Homeostasis through Peroxiredoxin-5. Nat Chem Biol 2019, 15 (12), 1232–1240. 10.1038/s41589-019-0399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Arrowsmith CH; Audia JE; Austin C; Baell J; Bennett J; Blagg J; Bountra C; Brennan PE; Brown PJ; Bunnage ME; Buser-Doepner C; Campbell RM; Carter AJ; Cohen P; Copeland RA; Cravatt B; Dahlin JL; Dhanak D; Edwards AM; Frederiksen M; Frye SV; Gray N; Grimshaw CE; Hepworth D; Howe T; Huber KVM; Jin J; Knapp S; Kotz JD; Kruger RG; Lowe D; Mader MM; Marsden B; Mueller-Fahrnow A; Müller S; O’Hagan RC; Overington JP; Owen DR; Rosenberg SH; Ross R; Roth B; Schapira M; Schreiber SL; Shoichet B; Sundström M; Superti-Furga G; Taunton J; Toledo-Sherman L; Walpole C; Walters MA; Willson TM; Workman P; Young RN; Zuercher WJ The Promise and Peril of Chemical Probes. Nature Chemical Biology 2015, 11 (8), 536–541. 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Sharma DK; Adams ST; Liebmann KL; Miller SC Rapid Access to a Broad Range of 6′-Substituted Firefly Luciferin Analogues Reveals Surprising Emitters and Inhibitors. Org. Lett 2017, 19 (21), 5836–5839. 10.1021/acs.orglett.7b02806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Sharma DK; Adams ST; Liebmann KL; Choi A; Miller SC Sulfonamides Are an Overlooked Class of Electron Donors in Luminogenic Luciferins and Fluorescent Dyes. Org. Lett 2019, 21 (6), 1641–1644. 10.1021/acs.orglett.9b00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Yao Z; Caldwell DR; Love AC; Kolbaba-Kartchner B; Mills JH; Schnermann MJ; Prescher JA Coumarin Luciferins and Mutant Luciferases for Robust Multi-Component Bioluminescence Imaging. Chem. Sci 2021, 12 (35), 11684–11691. 10.1039/D1SC03114G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Love AC; Caldwell DR; Kolbaba-Kartchner B; Townsend KM; Halbers LP; Yao Z; Brennan CK; Ivanic J; Hadjian T; Mills JH; Schnermann MJ; Prescher JA Red-Shifted Coumarin Luciferins for Improved Bioluminescence Imaging. J. Am. Chem. Soc 2023, 145 (6), 3335–3345. 10.1021/jacs.2c07220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Meinig JM; Ferrara SJ; Banerji T; Banerji T; Sanford-Crane HS; Bourdette D; Scanlan TS Targeting Fatty-Acid Amide Hydrolase with Prodrugs for CNS-Selective Therapy. ACS Chem. Neurosci. 2017, 8 (11), 2468–2476. 10.1021/acschemneuro.7b00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


