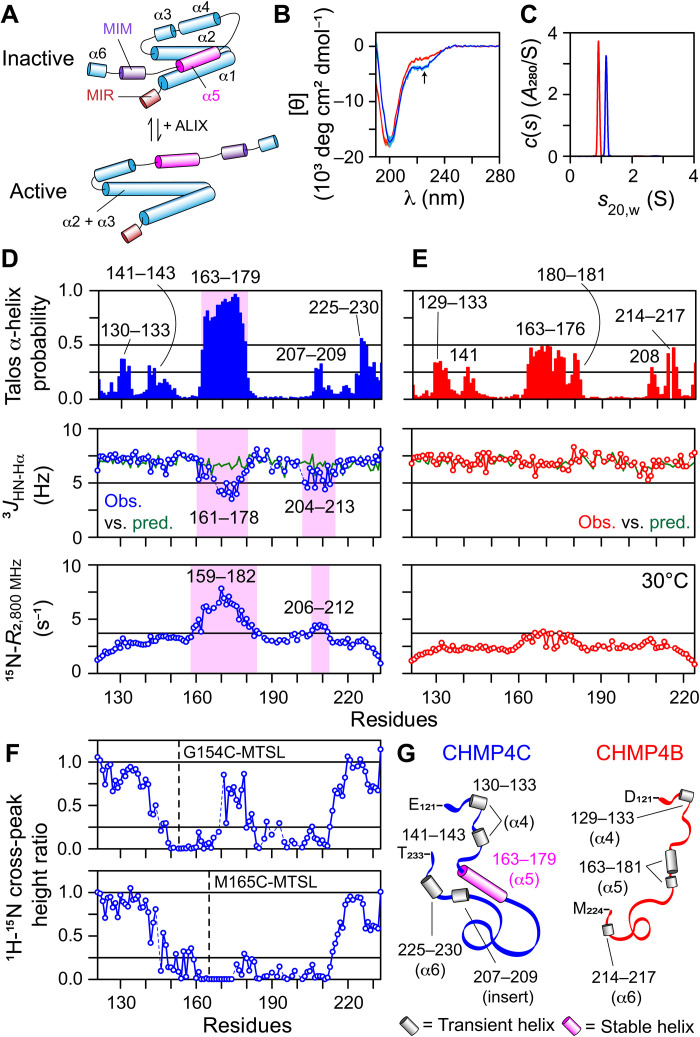
Fig. 5. Conformation and dynamics of CHMP4 paralogs in solution.
(A) Hypothetical models of CHMP4 (2, 4, 32, 33), originally described by Vietri et al. (2). MIM, MIT-interacting motif, which binds to the MIT domain containing proteins (e.g., VPS4) (72); MIR, membrane insertion region. Helix α5 (pink) is likely responsible for CHMP4 autoinhibition. (B) Far-UV CD spectra (five scans), mean (solid line) and SD (shaded region), and (C) sedimentation profiles of (blue) and (red). Arrow points to the dip at ~222 nm in the CD spectrum of . NMR analysis of (D) and (E) , including TALOS-N–derived helical propensities (top), a comparison between experimental 3JHN-Hα couplings against random coil values (middle), and 15N-R2 profiles measured at 30°C (table S1). Unlike , showed greater helical propensity for residues 163 to 179, highlighted by semitransparent magenta rectangle. The corresponding drop in 3JHN-Hα couplings and elevated 15N-R2 values of this region confirmed the presence of a stable helix. The insert region of (see Fig. 3B) exhibited deviations from random coil 3JHN-Hα couplings and elevated 15N-R2 values (highlighted by semitransparent magenta rectangles), indicating a residual helical structure. Residues of the regions showing elevated TALOS-N–derived helical propensities (ranging between 0.25 and 0.56), indicating transient helical structures, are labeled. (F) Evidence of transient long-range interactions in CHMP4C using experimental PRE profiles of (top) and (bottom). The locations of paramagnetic label, MTSL, are marked with dashed lines. (G) Schematic of (blue) and (red) based on NMR results. Transient and stable helices are depicted as gray and magenta cylinders, respectively; numbers denote the corresponding residues. The CHMP4 helices based on the proposed models of full-length proteins [see (A)] are in parentheses.