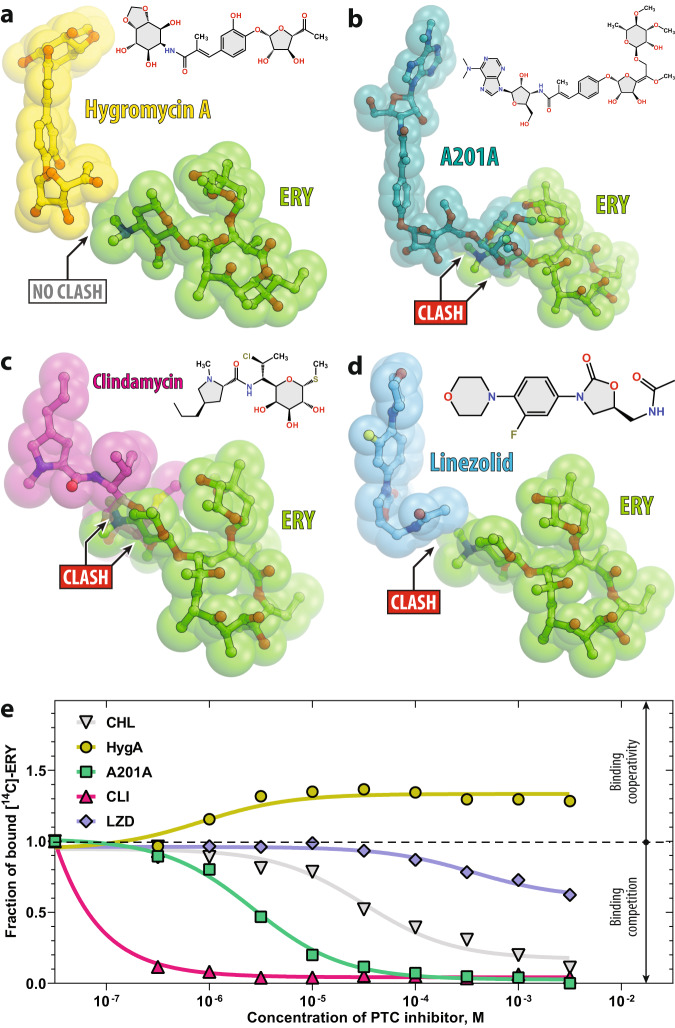
Fig. 1. Comparison of the macrolide binding site with those of various PTC-targeting antibiotics.
Superposition of the structures of the ribosome-bound ERY (red, PDB entry 6XHX20 [10.2210/pdb6XHX/pdb]) and a hygromycin A (yellow, PDB entry 5DOY34 [10.2210/pdb5DOY/pdb]), b nucleoside antibiotic A201A (teal, PDB entry 4Z3S34 [10.2210/pdb4Z3S/pdb]), c clindamycin (magenta, PDB entry 4V7V35 [10.2210/pdb4V7V/pdb]), or d linezolid (light blue, PDB entry 7S1G65 [10.2210/pdb7S1G/pdb]). All structures of ribosome-bound antibiotics were aligned based on domain V of the 23S rRNA. Note that only hygromycin A does not sterically clash with ribosome-bound ERY, while A201A, clindamycin, and linezolid overlap with the desosamine sugar of ERY. e Competition binding assay to assess the release of [14C]-radiolabeled ERY from the 70S ribosomes in the presence of increasing concentrations of one of the PTC-targeting antibiotics shown in (a)–(d). Chloramphenicol (CHL, gray) is used as a positive control known to compete with ERY37. The amount of [14C]-ERY associated with the ribosomes in the absence of a competitor drug is arbitrarily assigned as 1.0 (dashed line). Under experimental conditions, this corresponds to ~50% of the 70S ribosomes bound to [14C]-ERY. The measurements were repeated twice with similar results. Source data are provided as a Source Data file. Note that, at high concentrations, only HygA (yellow) stimulates additional binding of ERY to the 70S ribosome, whereas A201A (teal), clindamycin (magenta), linezolid (light blue), or chloramphenicol (gray) cause its dissociation.