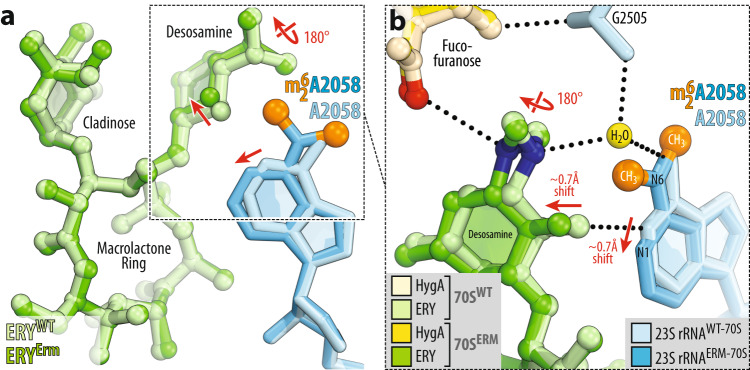
Fig. 8. Comparison of structures of erythromycin bound to the WT and Erm-modified 70S ribosome in the presence of hygromycin A.
a, b Superposition of ERY (light green) in complex with the WT 70S ribosome containing unmodified residue A2058 (light blue) and the structure of ERY (green) in complex with the Erm-modified 70S ribosome containing N6-dimethylated residue A2058 (blue with methyl groups highlighted in orange). Note that, while the overall position of ribosome-bound ERY is nearly identical in the two structures, N6-dimethylation of A2058 results in a 180-degree rotation of the dimethylamino group of desosamine away from A2058 nucleobase toward the fucofuranose moiety of HygA and formation of a direct H-bond with it. Also, note that N6-dimethylation of A2058 causes a small shift of ERY’s desosamine moiety away from the nucleotide, potentially weakening the H-bond between the 2’-OH of macrolide and the N1 atom of A2058 (red arrows).