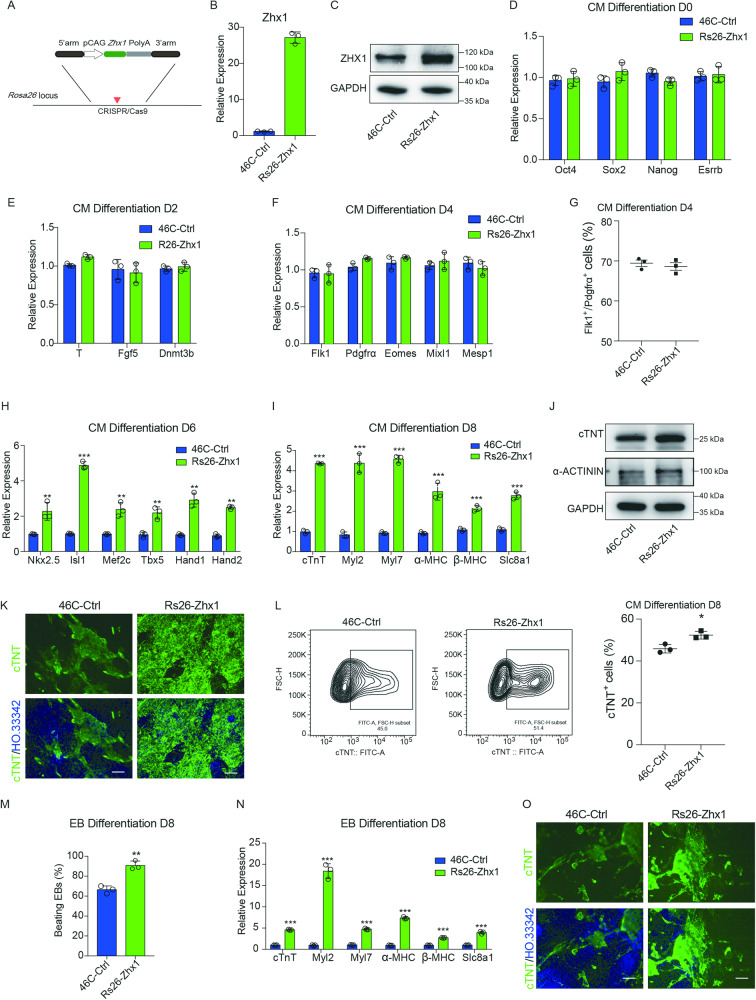
Fig. 3. Overexpression of Zhx1 promotes cardiac differentiation.
A The integrating strategy of the Zhx1 gene in the Rosa26 locus via CRISPR/Cas9 technology. B, C The mRNA (B) and protein levels (C) of Zhx1 overexpression. D The expression of pluripotent genes (Oct4, Sox2, Nanog, and Esrrb) after Zhx1 overexpression. E The expression of epiblast genes (T, Fgf5, and Dnmt3b) on day 2 of cardiomyocyte differentiation after Zhx1 overexpression. F The expression of mesoderm genes (Flk1, Pdgfrα, Eomes, Mixl1, and Mesp1) on day 4 of cardiomyocyte differentiation after Zhx1 overexpression. (G) The percentage of Flk1+/Pdgfrα+ cells on day 4 of cardiomyocyte differentiation after Zhx1 overexpression. H, I The expression of the CP genes (Nkx2.5, Isl1, Mef2c, Tbx5, Hand1, and Hand2) (H) and cardiomyocyte markers (cTnT, Myl2, Myl7, α-MHC, β-MHC, and Slc8a1) (I) after Zhx1 overexpression during cardiomyocyte differentiation. J The western blot results for the expression of cTNT and α-Actinin at day 8 of cardiomyocyte differentiation after Zhx1 overexpression. K The expression of cTNT protein after Zhx1 overexpression during cardiomyocyte differentiation through immunofluorescent staining. L The percentage of cTNT+ cardiomyocytes after Zhx1 overexpression. M The percentage of beating EBs on day 8 of EB differentiation after Zhx1 overexpression. N The expression of cardiomyocyte markers after Zhx1 overexpression on day 8 of EB differentiation. O The expression of cTNT protein after Zhx1 overexpression during the EB differentiation. Scale bar, 100 μm. Data are presented as the mean ± SEM (n = 3). The statistical significance is performed according to Student’s t-tests (unpaired two-tailed). *p < 0.05, **p < 0.01, and ***p < 0.001 versus 46C-Ctrl.