Fig. 5. Class A3 evasins achieve broad chemokine binding through flexible structure.
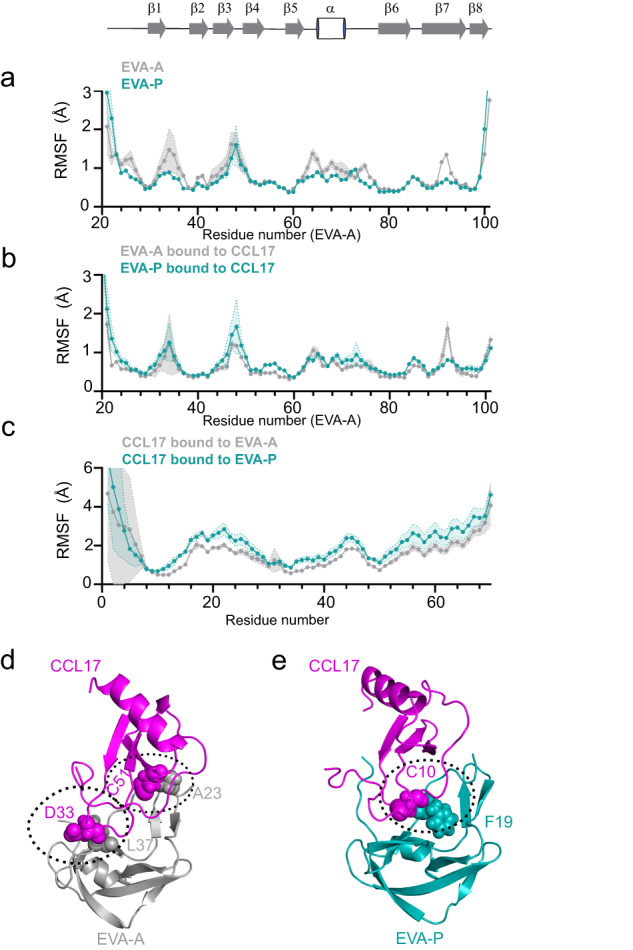
a, b Plots comparing the root-mean-square fluctuation (RMSF) values during MD simulations for EVA-A (grey) and EVA-P (green teal). The secondary structure of EVA-A in the crystal structure is shown at the top. a Free evasins only. b Bound state with chemokine. c RMSF of CCL17 bound to EVA-A (grey) and EVA-P (teal) with MD trajectories aligned at the evasins. Data are presented as mean ± SEM from three independent simulations. d The EVA-A:CCL17 complex forms high occupancy intermolecular hydrogen bonds at both ends of CC the motif. e The EVA-P:CCL17 complex forms a single high occupancy intermolecular hydrogen bond in the centre of the interface.
