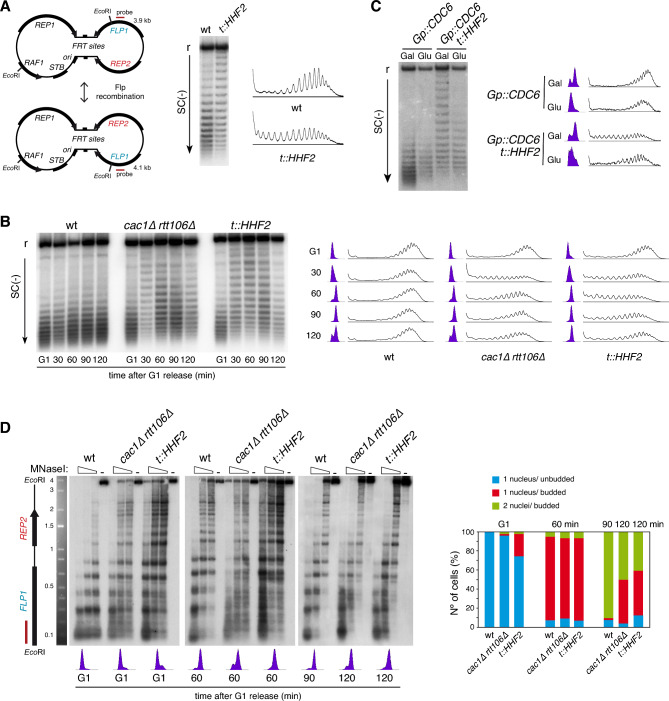
Figure 1.
Defective replication-coupled histone deposition causes transient changes in DNA topology and chromatin structure of the 2µ plasmid. (A) Plasmid topoisomer distribution of the 2µ plasmid in asynchronous cultures of wild type and t::HHF2 cells. A scheme of the two versions of the 2µ plasmid generated by Flp recombination, with the two unique halves and the intervening inverted repeat (FRT), is shown on the left. (B) Plasmid topoisomer distribution of the 2µ plasmid in wild type, cac1∆ rtt106∆ and t::HHF2 cells synchronized in G1 and released into fresh medium for different times. (C) Plasmid topoisomer distribution of the 2µ plasmid in Gp::CDC6 and Gp::CDC6 t::HHF2 cells synchronized in G1 and released into fresh medium in galactose or glucose-containing medium to express or not Cdc6, respectively. (A–C) Cell cycle progression and topoisomers profiles are shown. r and SC(−) indicate relaxed and negative supercoiling, respectively. Images show only the distribution of monomeric forms, as the higher-order forms reflect multimeric structures associated with the rolling circle replication mechanism of the 2µ plasmid 45, which are not a good readout to detect chromatin alterations (see supplementary Figures for complete and cropped images). (D) Chromatin structure of the EcoRI fragment spanning the FLP1 (bottom) and REP2 (top) genes from the 2µ plasmid in wild type, cac1∆ rtt106∆ and t::HHF2 cells synchronized in G1 and released into fresh medium for different times. See Fig. 1A for the position of the probe at the EcoRI fragment. Samples were run into different gels due to space limitations, and processed in parallel. Cell cycle stage of wild type, cac1∆ rtt106∆ and t::HHF2 cells was determined by FACS, cell morphology and DAPI (4′,6′-diamidino-2-phenylindole) staining of nuclei. (A–D) Original gels are presented in Fig. S1. The experiments were repeated at least twice with similar results.