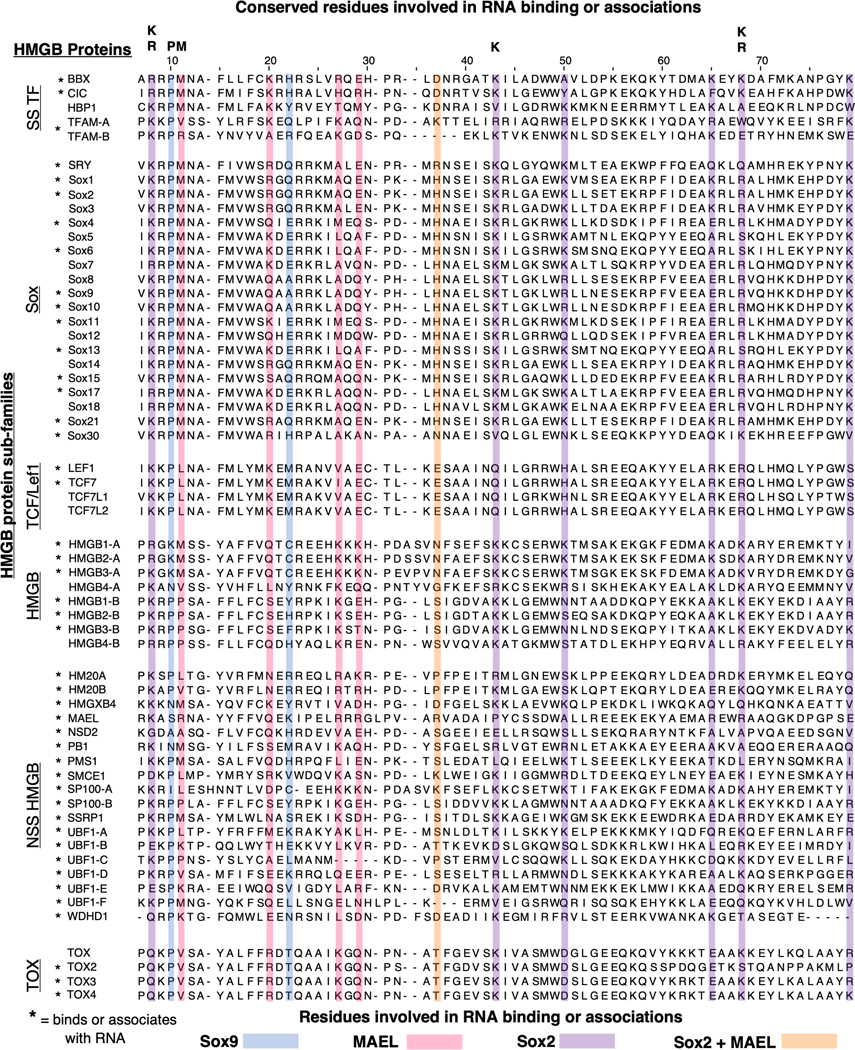
Figure 2.
HMGB-DBD sequence similarities suggests RNA binding potential. Sequence alignment, using MUSCLE,(Madeira et al., 2022) of the 48 human HMGB domains with sequences obtained from UniProt (UniProt, 2021) On the left, HMGBs are split into their respective subfamilies which have higher conservation within the subfamily (Sox, TCF/Lef1, HMGB, TOX), other HMGBs do not have a defined subfamily (non-sequence specific (NSS) and sequence specific (SS)). Sox9 (blue)(Girardot et al., 2018), MAEL (magenta)(Genzor & Bortvin, 2015), Sox2 (purple)(Holmes et al., 2020), Sox2 + MAEL overlap (orange) mutated residues are involved in RNA binding or associations. Some of these residues are highly conserved throughout the HMGB family (top). * = HMGB with evidence for binding or associating with RNA.