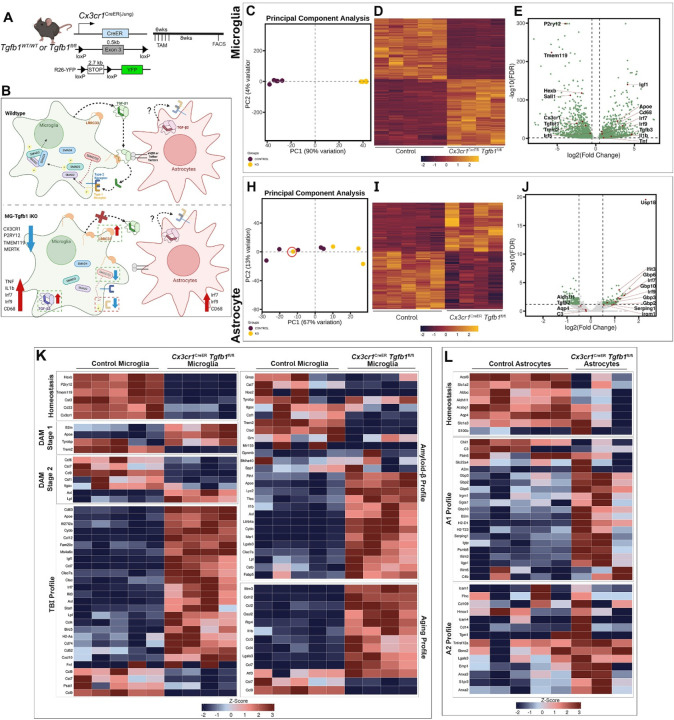
Figure 7. Transcriptomic analysis of microglia and astrocyte cells sorted from Cx3Cr1CreER(Jung)Tgfb1fl/fl mice.
(A) Mouse model used to induce Tgfb1 KO and YFP reporter in microglia. (B) Summary of transcriptomic changes in microglia or astrocytes pertaining to both inflammatory responses and critical TGF-β signaling component genes. (C, H) PCA analysis plot of microglia and astrocyte samples. Note that one astrocyte sample from iKO mice clustered irregularly in the PCA plot which has an RNA Integrity Number (RIN) below 8 (red circle). (D, I) Heatmap of expression of significantly differentially expressed genes in microglia and astrocytes from control and iKO samples. (E, J) Volcano plot showing expression log fold changes. (K) Microglial differential gene expression observed across various gene sets including, homeostatic microglia genes39–42, stage 1 and 2 disease-associated microglia (DAM) genes40, injury exposed microglial (TBI)42, amyloid beta exposed microglia39,41,42, and aged microglia41,42. (L) Astrocytic differential gene expression was observed across different gene sets including, homeostatic astrocyte genes, A1, and A2 genes. Z-scores were calculated and plotted to display differential gene expression43. The astrocyte sample that had an RIN< 8 was excluded from this analysis.