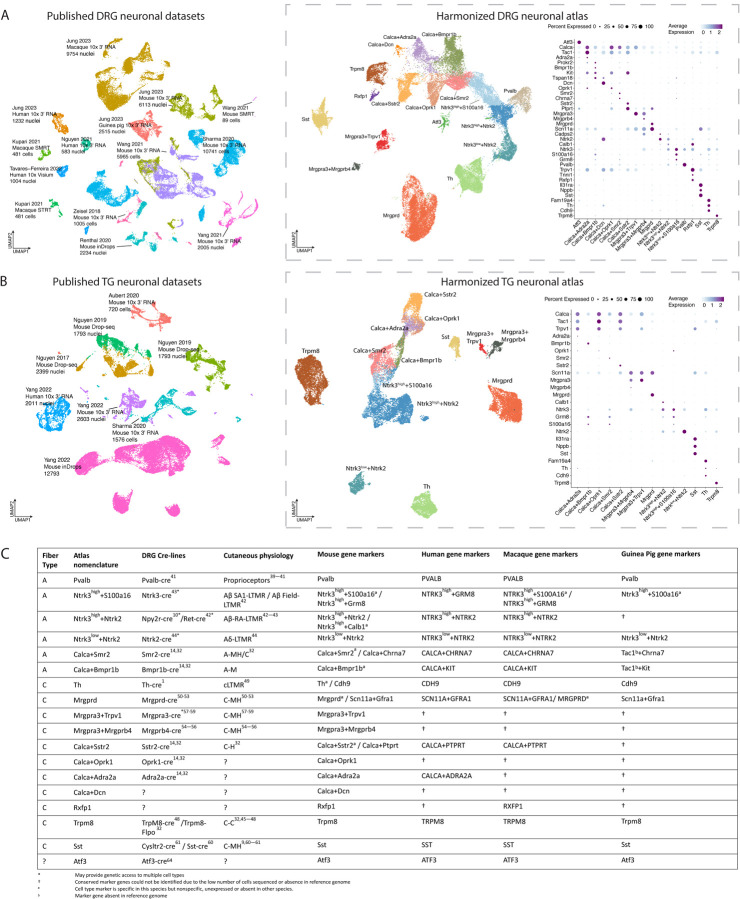
Figure 1: Integration of DRG or TG sc/snRNA-seq studies into harmonized neuronal atlases.
A. Integration of DRG neuronal sc/snRNA-seq datasets. Left: Co-clustering of the 9 sc/snRNA-seq studies used in the harmonized neuronal DRG atlas. Each study’s citation, sequencing technology, species, and number of cells/nuclei sequenced are listed. Cells/nuclei are colored by study. Middle: UMAP projection of harmonized DRG neuronal atlas (44,173 cells/nuclei). Cells/nuclei are colored by their final cell type annotations in the harmonized atlas, which are named with their defining marker genes. Right: Dot plot of cell-type-specific marker gene expression. Dot size indicates the fraction of cells/nuclei expressing each gene and color indicates average log-normalized scaled expression of each gene. Abbreviations: SMRT = single molecule real time sequencing; STRT = Single-Cell Tagged Reverse Transcription sequencing.
B. Integration of TG neuronal sc/snRNA-seq datasets. Left: Co-clustering of the 5 sc/snRNA-seq studies used in the harmonized neuronal TG atlas. Each study’s citation, sequencing technology, species, and number of cells/nuclei sequenced are listed. Cells/nuclei are colored by study. Right: UMAP projection of harmonized TG neuronal atlas (26,304 cells/nuclei). Cells/nuclei are colored by their final cell type annotations in the harmonized atlas. Right: Dot plot of cell-type-specific marker gene expression. Dot size indicates the fraction of cells/nuclei expressing each gene and color indicates average log-normalized scaled expression of each gene.
C. Somatosensory cell type nomenclature. Atlas nomenclature along with fiber type, corresponding Cre-recombinase line, cutaneous physiology, and cell-type marker genes for each species. Abbreviations: LTMR = low threshold mechanoreceptor; C-M = C-fiber responsive to mechanical stimuli; C-H = C-fiber responsive to hot stimuli; C-C = C-fiber responsive to cold stimuli; C-MH = fiber responsive to mechanical and heat stimuli; A-M = A-fiber responsive to mechanical stimuli; A-MH/C = responsive to mechanical, heat and cold stimuli.