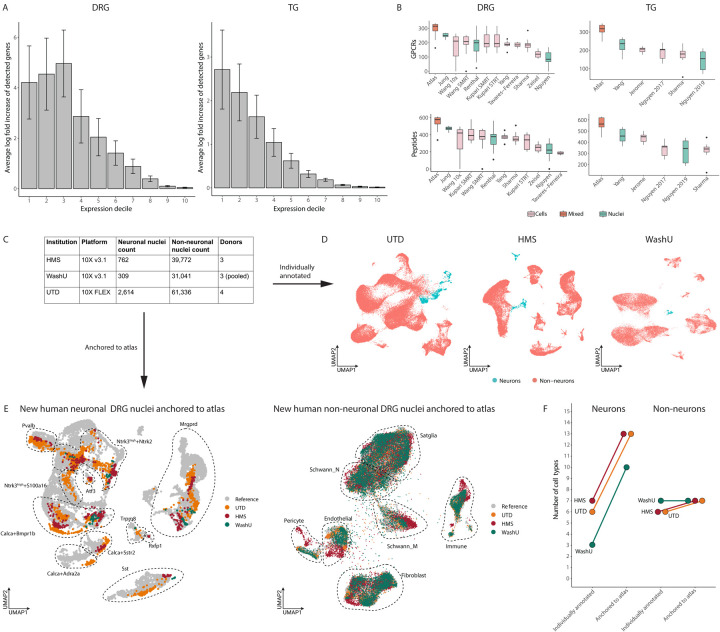
Figure 3: Harmonized reference atlas improves annotations of new human DRG snRNA-seq data.
A. Increased transcriptomic coverage of harmonized atlases compared to individual DRG and TG sc/snRNA-seq datasets. Bar plots display the fold increase between the average number of detected genes per cell type in the harmonized atlas and the average number of detected genes in the respective cell type of each individual DRG or TG sc/snRNA-seq dataset. Expression deciles are defined using the atlas counts matrix where bin 1 represents genes with lowest expression and bin 10 represents genes with the highest expression. Error bars represent standard deviation.
B. Increased transcriptomic coverage of GPCRs and peptides in harmonized atlases compared to DRG and TG sc/snRNA-seq datasets. Each box and whisker plot represents the distribution of the number of detected GPCRs or peptides per cell type in either the harmonized atlas or individual sc/snRNA-seq datasets. Studies with multiple species were combined. Dots represent outliers.
C. Number of nuclei sequenced in three new human DRG snRNA-seq datasets. Human DRGs from 10 donors were sequenced at three different sites: University of Texas-Dallas (UTD), Harvard Medical School (HMS) and Washington University at St. Louis (WashU).
D. Neurons and non-neurons of the new human DRG snRNA-seq datasets. UMAP visualization of 63,950 nuclei; WashU 31,350 nuclei; HMS 40,534 nuclei. Nuclei are colored by cell type.
E. Reference based cell type annotation of new human DRG snRNA-seq data. UMAP projection of new human neuronal (left) and non-neuronal (right) data colored by institution. Cell types annotations of new snRNA-seq data after anchoring to the harmonized reference atlas are circled. Only nuclei with an anchoring score of >0.5 are displayed. Non-neuronal projection was downsampled to 100,000 nuclei to improve visualization.
F. Neuronal subtype resolution after anchoring new human DRG snRNA-seq data to reference atlas. Plot of the number of DRG cell types that could be annotated individually compared to after anchoring to the reference atlas.