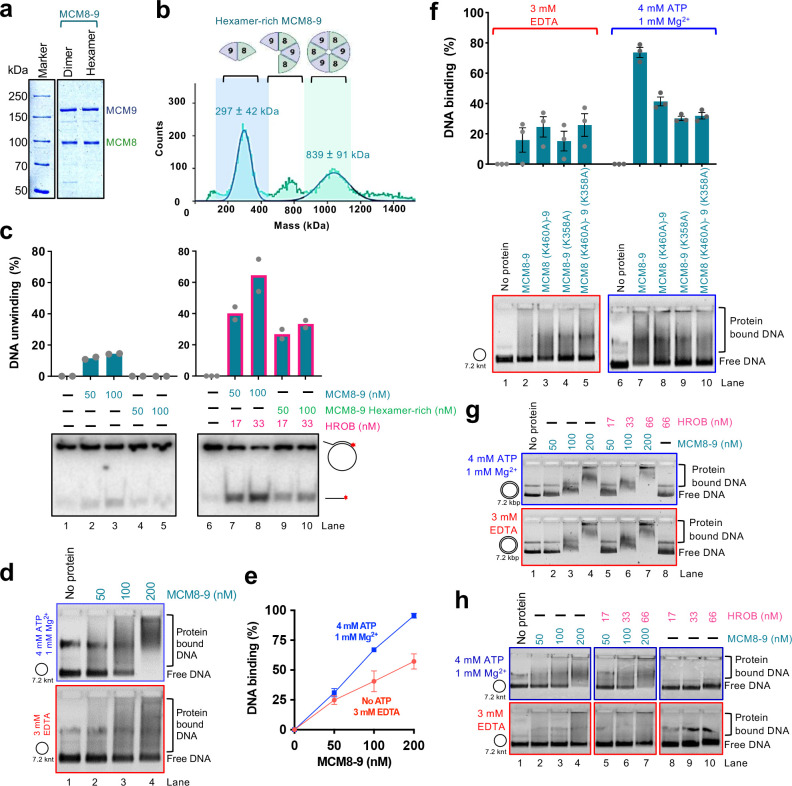
Fig. 5. ATP locks the MCM8–9 ring on ssDNA, irrespectively of HROB.
a. Purified wild type standard (dimer-rich) and hexamer-rich MCM8–9 preparations used in this study.
b. Mass photometry-based molecular weight distributions of hexamer-rich MCM8–9, purified by size exclusion chromatography. Compare with Fig. 4h. Error, SD.
c. DNA unwinding using standard (Fig. 4h) and hexamer-rich (Fig. 5b) preparations of MCM8–9, with and without HROB, as indicated using M13-based circular ssDNA substrate with 14 mM NaCl. The red asterisk indicates the position of the radioactive label. Top, quantification; Bars show range; n = 2; bottom, a representative experiment.
d. Electrophoretic mobility shift assays with human MCM8–9, with or without ATP, as indicated, using M13-based circular ssDNA substrate. A representative experiment is shown.
e. Quantification of assays such as shown in panel d. Error bars, SEM; n = 3.
f. Electrophoretic mobility shift assays with ATP-binding deficient variants of human MCM8–9 (100 nM), with or without ATP, as indicated, using circular M13-based ssDNA. Top, quantification; error bars, SEM; n = 3; bottom, a representative experiment.
g. Electrophoretic mobility shift assays with MCM8–9, with and without ATP, as indicated, using M13-based circular dsDNA as a substrate. Representative experiments are shown.
h. Electrophoretic mobility shift assays with human MCM8–9, with and without HROB, with and without ATP, as indicated, using circular M13-based ssDNA as a substrate. Representative experiments are shown.