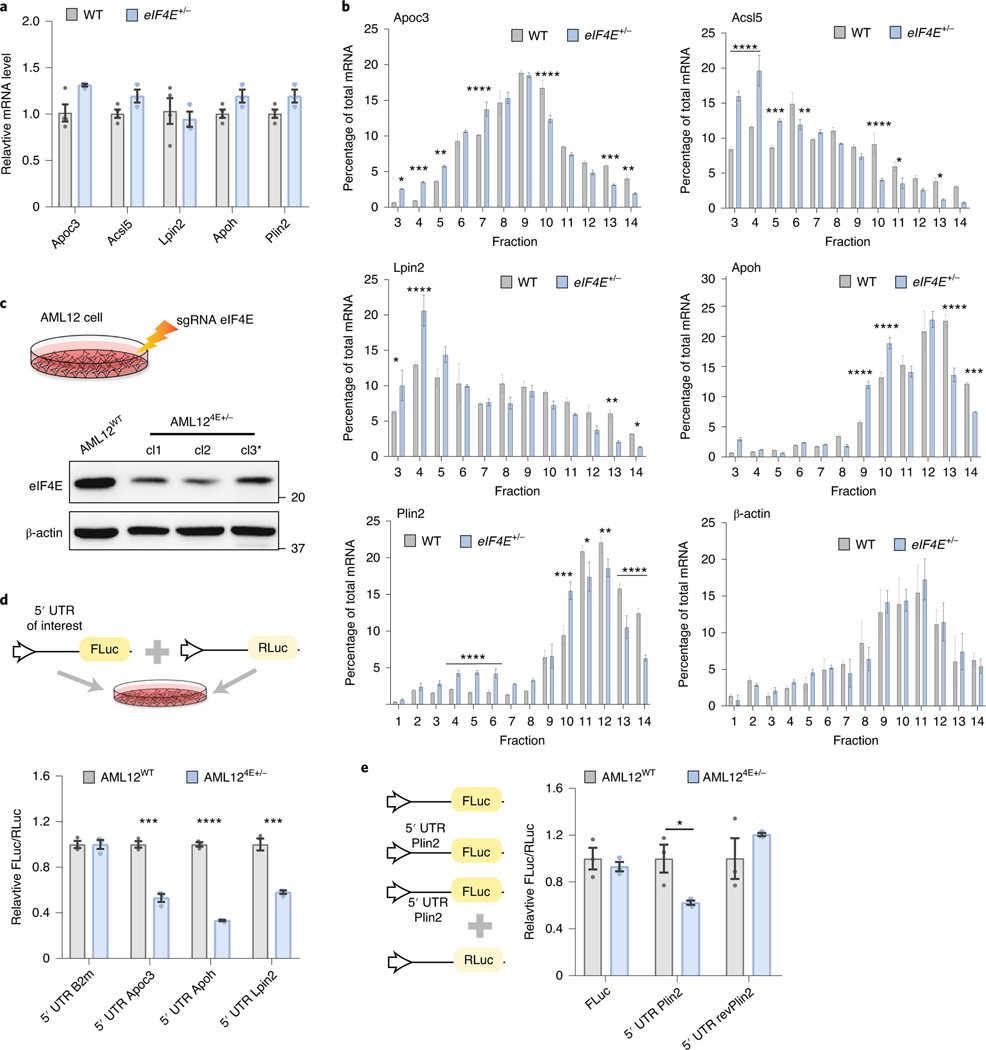
Fig. 3 |. Translational control through eIF4E regulates lipid-processing/storage proteins.
a, qPCR analysis of relative mRNA levels of labelled transcripts in WT or eIF4E+/− livers on HFD for 20 weeks; β-actin was used as internal control (n = 4, 3 independent biological replicates). b, Representative qPCR analysis of labelled genes isolated from sucrose gradient fractions of WT or eIF4E+/− livers on HFD. Values correspond to the percentage of total mRNA across fractions where free ribosomal subunits correspond to lower fractions (<5), 80 S monosome or low polysome fractions 6–10, and high polysomes within fractions 11–14 (n = 3, 3 technical replicates; mean ± s.e.m., *P < 0.05, **P < 0.01, ***P < 0.05, ****P < 0.0001, by unpaired, two-tailed Student’s t-test). c, Schematic depicting AML12 cell lines that were electroporated with CRISPR/Cas9 and sgRNA targeting Eif4e. Three independent clones are depicted below, with representative immunoblots. *Clone 3 (cl3) was used for subsequent experiments. d, Schematic depicting dual-luciferase assay, with target 5′ UTR of interest upstream of FLuc and RLuc included as reference control. Top: relative dual-luciferase expression in relation to 5′ UTR of interest within AML12 cell lines; bottom: n = 3, 3 independent replicates; mean ± s.e.m., ***P < 0.002, ****P < 0.00001 versus control by unpaired, two-tailed Student’s t-test. e, Relative dual-luciferase expression in relation to reporter depicted within AML12 cell lines (n = 3, 3 independent replicates; mean ± s.e.m., *P = 0.035 versus control by unpaired, two-tailed Student’s t-test).