Abstract
The paucity of blood granulocyte populations such as neutrophils in laboratory mice is a notable difference between this model organism and humans, but the cause of this species-specific difference is unclear. We previously demonstrated that laboratory mice released into a semi-natural environment, referred to as rewilding, display an increase in blood granulocytes that is associated with expansion of fungi in the gut microbiota. Here, we find that tonic signals from fungal colonization induces sustained granulopoiesis through a mechanism distinct from emergency granulopoiesis, leading to a prolonged expansion of circulating neutrophils that promotes immunity. Fungal colonization following either rewilding or oral inoculation of laboratory mice with Candida albicans induced persistent expansion of myeloid progenitors in the bone marrow. This increase in granulopoiesis conferred greater long-term protection from bloodstream infection by Gram-positive bacteria than by the trained immune response evoked by transient exposure to the fungal cell wall component β-glucan. Consequently, introducing fungi into laboratory mice may restore aspects of leukocyte development and provide a better model for humans and free-living mammals that are constantly exposed to environmental fungi.
One Sentence Summary:
Fungal colonization of lab mice, through release into the wild or inoculation with Candida albicans, enhances granulopoiesis.
INTRODUCTION
Laboratory mice are extensively utilized for studying how the microbiota contributes to the differentiation and function of the immune system (1-5). However, laboratory mice kept in ultra-hygienic specific pathogen-free (SPF) facilities lack the microbial exposure that humans and free-living mammals experience in a more complex environment. The artificial laboratory condition may give rise to differences between mice and humans in terms of proportions of leukocyte subsets, immune response to microbial challenges, and pathogenesis outcomes (6-8). Consistent with a role for microbes in this discrepancy, laboratory mice display a more mature immune system resembling that of an adult human following serial infection by pathogens or co-housing with ‘dirty’ mice harboring pathogens (9-12). In addition to lack of exposure to disease-causing pathogens, an artificial microbiota composition may also contribute to the lack of concordance between observations made in laboratory mice and humans. Mice reconstituted with microbiota from wild mice through fecal microbiome transplantation (FMT) or vertical transmission better recreate the unresponsiveness to immunotherapies that failed in clinical trials and display an altered course of influenza infection and colorectal tumorigenesis in these respective disease settings (13, 14).
The gut microbiota consists of multiple kingdoms of diverse unicellular organisms, and when the term is applied to agents transmitted through co-housing and FMTs, can include viruses and multicellular parasites with immunogenic properties. Thus, the respective contribution of symbiotic bacteria versus other microbial agents in the wild microbiota to immune development requires clarification. Another barrier towards investigating the natural microbiota is that wild mice harbor both environmental microbes and transmissible agents that straddle the line between commensal and pathogen. To address these issues, we recently established a mesocosm system termed ‘rewilding’ in which laboratory mice experience a semi-natural environment through release into an outdoor enclosure facility (15). A key feature of this outdoor enclosure facility is that the fencing system prevents contact with wild rodents that potentially harbor pathogens while allowing the mice to freely explore and encounter the natural plant and insect fauna. As such, rewilded laboratory mice are exposed to environmental microbes but remain seronegative for pathogens excluded from SPF facilities (16). We previously demonstrated that rewilded mice display hallmarks of increased immune activation compared with matched control laboratory mice, which included a striking expansion of granulocytes, a group of myeloid lineage white blood cells that include first responders such as neutrophils (16, 17). The gut microbiota of rewilded mice displayed >100-fold increases in fungal burden compared with laboratory mice, and transferring fungi isolated from feces of rewilded mice into laboratory mice reproduced the increase in peripheral granulocytes. We were able to mimic this effect by colonizing the intestines of laboratory mice with Candida albicans, a model fungus that is commonly detected in the human gut.
C. albicans, a gut commensal and opportunistic pathogen, colonizes most individuals since childhood (18-20). The transition from yeast to hyphal cell morphology has been shown to regulate the balance between commensalism and invasive infection in the gastrointestinal tract (21-27). Commensal C. albicans can stimulate the differentiation of CD4+ T helper 17 (Th17) cells to improve immunity, fortify the intestinal barrier, and regulate social behavior (28-33). In addition to this adaptive immune response mediated by lymphocytes during gut colonization, bloodstream C. albicans infection can prime an innate immune memory response mediated by the myeloid compartment termed trained immunity. To meet the high demand for phagocytes to contain invasive pathogens, progenitors in the bone marrow (BM) rapidly ramp up the differentiation of myeloid cells through the process of emergency granulopoiesis, which returns to baseline levels following resolution of the infection (34-36). However, a number of studies showed the response to C. albicans or fungal products has long-term consequences due to the reprogramming of the myeloid compartment, resulting in an enhanced response to reinfection (35, 37-42). In this setting, β-glucan, a universal-fungal cell wall component, activates Dectin-1 and the signaling adaptor CARD9 to alter chromatin accessibility in trained monocytes and macrophages, which enhances production of inflammatory cytokines (41, 43, 44). Although Th17 differentiation following local activation of CX3CR1+ mononuclear phagocytes during intestinal colonization by C. albicans is also dependent on Dectin-1 (45), it is unclear whether blood granulocyte expansion in rewilded mice involves this pathway. Similarly, the qualitative differences between mobilization of granulocytes following gut colonization versus transient bloodstream exposure to fungi require a better understanding.
Here, we found that C. albicans inoculation and fungal colonization following rewilding of laboratory mice increased the number and frequency of multipotent progenitors (MPPs) and the myeloid-biased MPP3 subset in the BM. Optimal expansion of MPP3s required IL-6 but were surprisingly independent of Dectin-1 and Th17 pathways. This enhanced granulopoiesis led to an increase in circulating CD62LhiCXCR4lo neutrophils in C. albicans colonized laboratory mice and rewilded mice, as well as increases in eosinophils exclusively in rewilded mice. A yeast-locked C. albicans mutant was unable to induce MPP3s in the BM and neutrophils in the blood, which was attributed to the lack of production of the hypha-associated factor candidalysin. In contrast to transient β-glucan-induced granulopoiesis, fungal colonization sustained the increased granulopoiesis, which was reflected by persistently high levels of peripheral neutrophils. Consequently, C. albicans colonized mice displayed prolonged protection against Staphylococcus aureus bloodstream infection and yielded greater immunity at later time points when compared side-by-side to the previously described β-glucan-induced trained immunity model. Taken together, these findings implicate intestinal fungi in continuously remodeling myeloid cell development in the BM and suggest that environmental fungi may, in part, correct the differences between laboratory mice and humans.
RESULTS
Intestinal C. albicans colonization enhances granulopoiesis in the bone marrow
We previously found that inoculating germ-free mice or antibiotics-treated laboratory mice with C. albicans (SC5314) mimics the increase in blood granulocytes observed in rewilded mice that are exposed to fungi, such as Aspergillus spp., in a more natural environment. C. albicans is a genetically tractable model fungus with isogenic mutants available for study, and its use allowed us to compare our findings with the literature on fungal colonization of the gut. Therefore, we used rewilding and C. albicans colonization models interchangeably in this study to cross-validate key observations regarding the mechanism of granulocyte expansion. Although we previously established that the increase in blood granulocytes in the antibiotics pretreatment model of C. albicans colonization was due to an increase in neutrophils (CD11b+Ly6G+), we did not examine additional cell surface markers that inform neutrophil biology. Surface levels of the chemokine receptors CXCR4 and cell adhesion molecule CD62L increase and decrease, respectively, as neutrophils age (46, 47). We found that the ratio of the newly released population (CD62LhiCXCR4lo) of neutrophils to the aged population (CD62LloCXCR4hi) of neutrophils substantially increased in antibiotics-treated C. albicans colonized mice compared with non-colonized control mice, suggesting that fungal colonization increases the number of fresh neutrophils in the blood (Fig. 1A and fig. S1A). These results raise the possibility that C. albicans enhances the production of neutrophils.
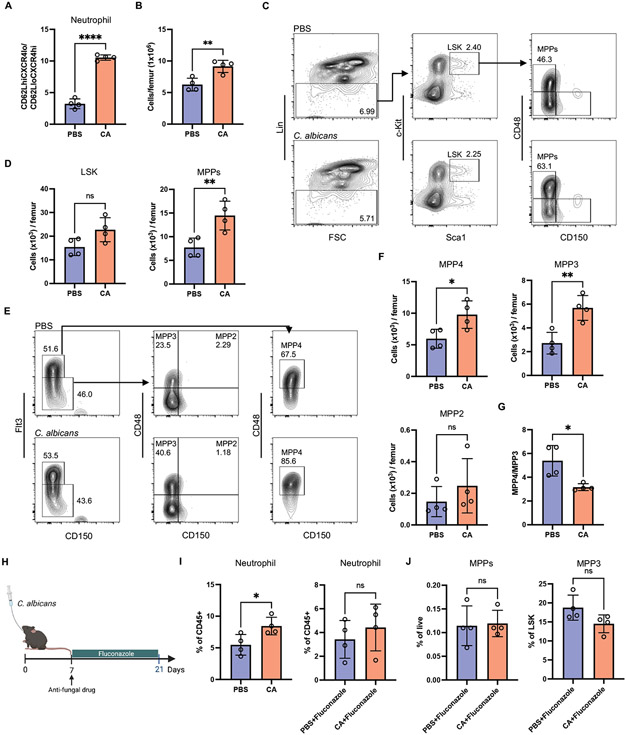
Fig. 1. Inoculation of laboratory mice with C. albicans drives expansion of myeloid progenitors.
(A) Ratio of newly released (CD62LhiCXCR4lo) population to aged (CD62LloCXCR4hi) population of Ly6G+ peripheral neutrophils from antibiotic-treated mice 21 days post-inoculation with PBS or C. albicans. N = 4 mice per group. (B) The cellularity of total BM cells from germ-free mice 21 days post-inoculation with PBS or C. albicans. N = 4 mice per group. (C) Representative flow cytometry plots depicting gating strategy for hematopoietic stem cells and progenitors in BM from germ-free mice inoculated with PBS or C. albicans. LSK cells were gated from Lin− cells and characterized as Sca1+c-Kit+ cells. A subpopulation of LSK was further characterized as MPP (CD48+CD150−LSK). (D) Quantification of number of LSK and MPPs from (C). N = 4 mice per group. (E) Representative flow cytometry plots depicting gating strategy used to identify MPP subsets in BM from germ-free mice inoculated with PBS or C. albicans. MPP4 cells were gated on Flt3+LSK and identified by CD48 and CD150. MPP3 and MPP2 cells were gated on Flt3−LSK and identified by CD48 and CD150. (F) Quantification of the number of MPP subsets from (E). (G) Ratio of MPP4 to MPP3. (H) Germ-free mice were administrated fluconazole in the drinking water continuously starting from day 7 post-inoculation with PBS or C. albicans. Blood and BM analysis were performed day 21 post-inoculation N = 4 mice per group. (I) Frequency of neutrophils in the peripheral blood of germ-free mice treated as in (H). (J) Frequency of MPPs and MPP3 in the BM of germ-free mice treated as in (H). Dots in bar graphs correspond to individual mice. Mean and SD are shown. *p < 0.05, **p < 0.01, ****p < 0.0001 by two-tailed Student’s t test between groups. ns, not significant.
Hematopoietic stem cells (HSCs) are defined as the Lin−Sca-1+c-Kit+ (LSK) fraction of the BM that give rise to non-self-renewing multipotent progenitors (MPPs). MPPs, comprised of three subsets, give rise to leukocyte populations. MPP2 and MPP3 are myeloid-biased MPP subsets that develop toward erythrocyte/megakaryocyte and monocyte/granulocyte lineages, respectively, while MPP4 commits to lymphoid lineage development (48, 49). To examine whether the expansion of peripheral neutrophils is attributed to the response of the myeloid progenitor pool, we used flow cytometry to analyze the BM in germ-free mice and antibiotics-treated mice colonized by C. albicans. Compared with mock-treated controls, germ-free mice 21 days after oral gavage with C. albicans displayed increases in the total cellularity of the femoral BM and the number of MPPs (CD48+CD150−LSK) but not HSCs (Fig. 1, B to D and fig. S1B). Additionally, the number of myeloid-biased MPP3 subset (Flt3−CD150−CD48+LSK) that gives rise to neutrophils was greatly increased (Fig. 1, E and F). Although absolute number of lymphocyte precursor MPP4s (Flt3−CD150−CD48+LSK) increased, the ratio of MPP4 to MPP3 was significantly reduced (Fig. 1G), indicating a skewing towards granulocyte differentiation. C. albicans colonized antibiotic-treated mice also displayed a comparable increase in total MPPs and the MPP3 subset but not MPP4 (fig. S1, C and D), demonstrating that fungal colonization favors myeloid-lineage commitment in both the germ-free and antibiotics pretreatment models. Treatment of C. albicans mono-associated mice with the antifungal drug fluconazole decreased fungal burden and reduced neutrophils in the blood along with the frequency of myeloid progenitor pools (Fig. 1, H to J and fig. S1E). These results suggest that persistent colonization with fungi is essential to maintain the elevated granulopoiesis.
Rewilding increases granulopoiesis
To determine whether granulopoiesis is enhanced following exposure to the natural environment, we performed an independent rewilding experiment in which 6-8 week old SPF laboratory mice were released into the enclosure and captured 6 weeks later, at which point their immune profile was compared to littermate controls kept in the conventional animal facility. Given that the previous rewilding experiment was performed in 2017, we first confirmed that granulocyte expansion was reproducible and not subject to year-to-year variation in the local environment of the outdoor facility (e.g., vegetation, weather, etc.). Despite the two-year interval between experiments, rewilded mice displayed a similar expansion of granulocytes (SSChi) in the periphery as before (Fig. 2A). Our previous rewilding experiment did not include flow cytometry markers for distinguishing granulocyte subsets. Although the increase in blood neutrophils was not statistically significant, a positive correlation between the abundance of circulating neutrophils and fungal burden was still observed in rewilded mice (fig. S2A). Additionally, neutrophils from rewilded mice displayed a dramatically altered cell surface staining for CD62L and CXCR4 compared with laboratory controls indicative of an increase in newly generated (CD62LhiCXCR4lo) cells (Fig. 2, B and C), similar to C. albicans colonized laboratory mice. Interestingly, rewilded mice displayed a substantial increase in eosinophils that was not observed in C. albicans colonized laboratory mice (Fig. 2A, S2B). This difference could be explained by the presence of other fungal taxa in the outdoor enclosure. To test this possibility, we gavaged laboratory mice every other day repeatedly for 2 weeks with a consortium of “wild” fungi that we previously isolated from rewilded mice (16), and found that this treatment induced an increase in peripheral eosinophils (fig. S2C). Thus, we believe C. albicans is useful for mechanistic probing how fungi induce neutrophil expansion, while wild fungi might better recapitulate the full effect of rewilding. Consistent with our previous finding that rewilded mice harbored a significant increase in stool fungal burden, we readily observed the presence of fungi associated with the mucus layer in the small intestine of rewilded mice. In contrast, fungi were nearly undetectable in laboratory mice (Fig. 2D). These results highlight the reproducibility of granulocyte expansion and fungal colonization in our rewilding system, and also identify eosinophils as a uniquely expanded population in mice captured from outdoors.
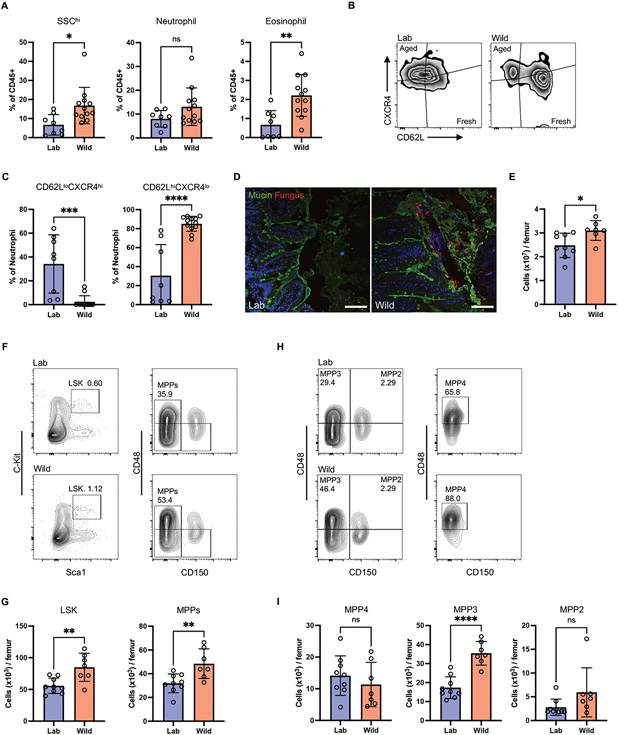
Fig. 2. Rewilding increases granulopoiesis.
(A) Frequency of granulocytes (SSChi), neutrophils (Ly6G+), and eosinophils (Siglec-F+) in the peripheral blood of rewilded mice (Wild) and control mice maintained in the laboratory condition (Lab). Neutrophils and eosinophils were gated on Live+CD45+CD11b+. N=8 lab and 12 rewilded mice. (B) Representative flow cytometry plots of newly released (CD62LhiCXCR4lo) and aged (CD62LloCXCR4hi) neutrophils. (C) Quantification of proportion of fresh and aged neutrophils. N=8 lab and 12 rewilded mice. (D) Confocal images of ileum sections immunostained with anti-candida antibody (non-specifically labels fungi) and counterstained with FITC-lectins mixture, which binds to oligosaccharide structures of mucins. Scale bars represent 100 μm. (E) The cellularity of total BM cells from lab and rewilded mice. N=9 lab and 7 rewilded mice (E, G, I). (F) Representative flow cytometry plots of LSK (Lin−cKit+Sca1+) and MPPs (CD48+CD150−LSK). (G) Cell number of LSK and MPPs in the BM of lab and rewilded mice. (H) Representative flow cytometry plots of MPP4, MPP3, and MPP2. (I) Cell number of MPP subsets in the BM of lab and rewilded mice. Dots in bar graphs correspond to individual mice. Mean and SD are shown. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by two-tailed Student’s t test between groups. ns, not significant.
Like C. albicans colonized mice, rewilded mice showed a significantly increased BM cellularity and the number and frequency of MPPs. Additionally, rewilded mice displayed a higher absolute number but not proportion of HSCs than laboratory mice (Fig. 2, E to G and fig. S2D). Rewilding also led to an increase in the number and frequency of myeloid-biased MPP3s and not lymphoid-biased MPP4s (Fig. 2, H to I and fig. S2E). In contrast to the periphery where the neutrophils are mature (Ly6G+), neutrophils in the BM can be segregated into mature Ly6G+CXCR2+ and immature Ly6Glo/+CXCR2− populations (50). Using these markers, we found the frequency of immature neutrophils increased and mature neutrophils decreased in the BM of rewilded mice compared to laboratory mice (fig. S2F). In human BM, the majority of neutrophils are immature. Therefore, we believe that fungal colonization brings the steady state of neutrophil development in laboratory mice to more closely resemble human BM development. Overall, these results demonstrate that rewilding leads to enhanced granulopoiesis giving rise to younger neutrophils in the periphery, similar to inoculation of laboratory mice with C. albicans.
Fungal colonization of the gut induces sustained changes to the host immune system
To determine whether the time course of granulopoiesis differs between invasive infection and gut colonization, we attempted to compare the immune response between oral and intravenous inoculation with C. albicans. Because the gut colonization model involves antibiotic pretreatment, mice received the same antibiotics regimen prior to intravenous injection with a non-lethal dose of C. albicans. Unexpectedly, intravenous injection led to similar levels of C. albicans gut colonization as the oral route of inoculation (Fig. 3A). Thus, we concluded that the intravenous C. albicans injection model is inappropriate for investigating transient exposure to fungi, and turned to a simpler established model of acute anti-fungal responses.
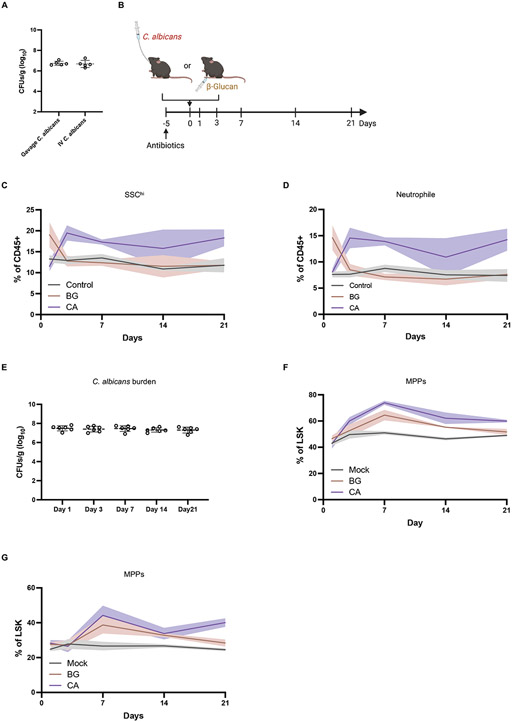
Fig. 3. Intestinal C. albicans colonization leads to sustained granulopoiesis.
(A) Colony-forming units (CFUs) of C. albicans in feces from antibiotic treated mice 21 days post-oral inoculation with C. albicans or intravenous injection with a non-lethal dose (1x104 CFUs). N = 5 mice per group. (B) Experimental model of C. albicans colonization and β-glucan injection in antibiotic-treated mice. Blood and bone marrow were collected on day 1, 3, 14, and 21 of inoculation for flow cytometry analysis. N=6 mice per group. (C and D) Frequency of granulocytes (SSChi) and neutrophils in the peripheral blood from mock, C. albicans colonized and β-glucan injected mice. (E) C. albicans CFUs in feces collected from antibiotic-treated mice inoculated with C. albicans on indicated days. (F and G) Frequency of MPPs and MPP3 in the bone marrow from mock, C. albicans-colonized and β-glucan injected mice.
β-glucan is an abundant fungal cell wall polysaccharide that elicits host responses through activating innate immune receptors such as Dectin-1 (51, 52). Therefore, we performed a time course analysis comparing the effect of intraperitoneal injection with β-glucan and oral gavage with C. albicans in antibiotics-treated mice. Intraperitoneal injection of β-glucan led to expansion of SSChi granulocytes and neutrophils in the blood as early as 24 hr post-treatment compared with control mice, which returned to baseline levels by day 3 (Fig. 3, B to D). In contrast, the frequency of peripheral granulocytes and neutrophils reached a plateau at day 3 in mice orally inoculated with C. albicans and remained high throughout the course of the experiment, matching the stable levels of colonization observed in the stool (Fig. 3, C to E). The number of blood eosinophils and monocytes were largely unaffected by β-glucan injection and C. albicans gut colonization (fig. S3, A and B). MPPs and MPP3s in the BM were increased and remained high in mice orally inoculated with C. albicans. However, different from the time course of neutrophil expansion in the blood, the increase in these progenitor populations following β-glucan injection led to a gradual decline rather than a steep drop at day 3 (Fig. 3, F and G). These findings indicate that fungal colonization of the gut leads to sustained granulopoiesis and stable expansion of neutrophils, while a single intraperitoneal injection of β-glucan induces transient changes.
Fungal colonization enhances granulopoiesis independently of dectin-1 and Th17 pathways but in a manner dependent on IL-6R signaling
Antigen presenting cells activated by Dectin-1 and other pattern recognition receptors that signal through CARD9 can induce the differentiation of Th17 cells that mediate neutrophil mobilization through IL-17A (53, 54). Therefore, it is possible that C. albicans colonization induces sustained Dectin-1 signaling. However, daily intraperitoneal injection of β-glucans for 7 days did not induce a significant increase in peripheral neutrophils and eosinophils (fig. S4A). Although repetitive β-glucan injection induced a slight increase in MPP3s, in contrast to C. albicans colonized mice, MPP2s were also increased and the ratio of MPP4 to MPP3 was comparable to the mock treated mice (fig. S4. B and C). These results raise the possibility that the effects of intestinal fungal colonization are Dectin-1 and CARD9 independent. Indeed, C. albicans colonization still induced increases in MPP3s in the BM and peripheral neutrophils in mice lacking Dectin-1 (Clec7a) or CARD9 (Fig. 4A and fig. S4D). Dectin-1- and CARD9-deficient mice displayed similar amounts of C. albicans in the stool as their wild-type littermates (Fig. 4B). Also, rewilded Dectin-1- and CARD9-deficient mice exhibited enhanced granulopoiesis, including an increased number in MPPs and MPP3, upon being released into the outdoor enclosure (Fig. 4C and fig. S4E). In addition to Dectin-1, we also examined the role of Toll-like receptor 2 (TLR2), which recognizes phospho-lipomannans and chitin from the fungal cell wall (55, 56). We found that C. albicans colonization induced increases in MPP3 in the BM and peripheral neutrophils in mice lacking TLR2 (fig. S4F). Similarly, in our previous study (16), rewilded mice deficient in NOD2, another molecule shown to sense fungi (57), exhibited a comparable expansion of granulocytes as rewilded wild-type mice.
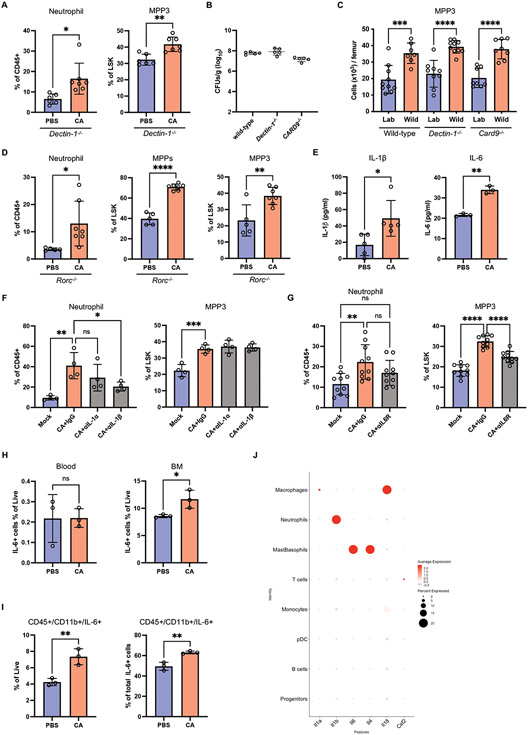
Fig. 4. Fungal colonization enhances granulopoiesis in a manner dependent on IL-6R signaling.
(A) Frequency of neutrophils in the peripheral blood and MPP3 in the BM from antibiotic-treated Dectin-1−/− mice 21 days post-inoculation with C. albicans. N = 6 PBS control and N = 7 C. albicans-colonized mice. (B) Fungal CFUs in feces from C. albicans-colonized wild-type, Dectin-1−/−, and Card9−/− mice on day 21. N = 5 mice per group. (C) Quantification of number of MPP3 in the BM of laboratory and rewilded wild-type (N = 9 lab and 7 rewilded mice), Dectin-1−/− (N = 8 lab and 10 rewilded mice), and Card9−/− (N = 8 lab and 8 rewilded mice) mice. (D) Frequency of neutrophils in the peripheral blood and MPPs and MPP3 in the BM from antibiotic-treated Rorc−/− mice 21 days post-inoculation with C. albicans. N = 5 PBS control and N = 7 C. albicans-colonized mice. (E) Quantification of IL-1β (N = 5) and IL-6 (N = 3) in the BM extracellular fluid from antibiotic-treated mice 21 days post-inoculation with PBS or C. albicans. (F) Frequency of neutrophils in the peripheral blood and MPP3 in the BM on day 7 from mice treated with anti-IL-1α, anti-IL-1β or IgG isotype control antibodies on day −1, 1, 3, and 5 days post-inoculation with C. albicans. N=4 mice per group. (G) Frequency of neutrophils in the peripheral blood and MPP3 in the BM from mice treated with anti-IL-6R or IgG isotype control antibodies on day −1, 1, 3, and 5 days post-inoculation with C. albicans. N = 10 mice per group. (H) Quantification of IL-6+ cells gated on live cells in the peripheral blood and BM from antibiotic-treated mice 21 days post-inoculation with PBS or C. albicans by flow cytometry. (I) Proportion of live IL-6+ cells that are CD45+CD11b+ (left panel) and quantification of live cells that are IL-6+ myeloid cells (CD45+CD11b+) (right panel) in the BM from antibiotic-treated mice 21 days post-inoculation with PBS or C. albicans. (J) Cytokine gene expression across cell types identified by scRNA-Seq analysis. Dots in bar graphs correspond to individual mice. Mean and SD are shown. *p < 0.05, **p < 0.01, ****p < 0.0001 by two-tailed Student’s t test between groups (A, D, E, H) and ordinary one-way analysis of variance (ANOVA) with Holm-Sidak multiple comparisons test (C, F-G). ns, not significant. Antibiotic-treated groups (A, D-I).
IL-17A can promote granulopoiesis by inducing increased circulating levels of granulocyte-colony stimulating factor (G-CSF) (58, 59). Therefore, we examined the possibility that the Th17 response mediates neutrophil expansion during C. albicans gut colonization. As demonstrated by others (33, 45), we observed that C. albicans colonization increased IL-17 production by CD4 T cells and the proportion of RoRγt+ CD4 T cells in the small intestine and colon (fig. S4, G and H). However, antibiotics-treated Rorc−/− mice, which are deficient in IL-17-producing cells, still displayed an in increase in blood neutrophils and expansion of MPPs and MPP3 in the BM upon C. albicans colonization (Fig. 4D). We also found that CD4 T cells in general were dispensable by depleting the CD4 T cells in C. albicans colonized wild-type mice (fig. S4I). Altogether, these results suggest that C. albicans colonization modulates neutrophil development independent of canonical fungal sensing receptors such as Dectin-1 and anti-fungal immune pathways.
Other cytokines are known to regulate the expansion of HSCs and differentiation into myeloid progenitors, such as IL-1, IL-6 and granulocyte macrophage colony-stimulating factor (GM-CSF) (60). As expected, we found that IL-1β and IL-6 were significantly increased in the BM of C. albicans colonized mice (Fig. 4E). We injected IL-1α or IL-1β-specific neutralizing antibodies to assess their contribution. Interestingly, C. albicans colonized mice undergoing IL-1α or IL-1β blockade displayed equivalent MPP3 levels to isotype control treated mice; yet, peripheral neutrophils were notably decreased in the anti-IL-1β injected mice (Fig. 4F). In contrast, blocking the IL-6 receptor (IL-6R) reduced the C. albicans-mediated increase in MPP3. The neutrophil levels of anti-IL6R-treated C. albicans colonized mice and control mice not colonized by C. albicans were similar (Fig. 4G). GM-CSF blockade did not reduce either neutrophils or MPP3 in C. albicans colonized mice (fig. S4J). Therefore, fungal colonization in the gut leading to an increase in granulopoiesis was dependent on IL-6R signaling and independent of IL-1 and GM-CSF. However, IL-1β might play a role in regulating neutrophil mobilization from the BM.
To test whether the increase in IL-6 upon C. albicans colonization was a result of local expansion of IL-6 producing cells, we harvested the blood and BM cells from mock-treated or C. albicans colonized mice and quantified IL-6+ cells by flow cytometry. C. albicans colonization increased the proportion of IL-6+ cells locally in the BM and not in the blood (Fig. 4H). IL-6+ myeloid cells (CD45+B220−CD3e−CD19+) increased in C. albicans colonized mice and were the main source of IL-6 in the BM (Fig. 4I and fig. S5A). To further define the cell types that contributed to Il6 expression, we performed single-cell RNA sequencing (scRNA-seq) on BM cells isolated from mock-treated and C. albicans colonized mice. We employed unsupervised dimensionality reduction analysis combined with unbiased cell type recognition using the ImmGenData open-source (expression) reference database to identify various hematopoietic populations consisting of murine progenitors, neutrophils, monocytes, plasmacytoid dendritic cells (pDCs), macrophages, Mast/basophils, and B and T lymphocytes (fig. S5B). The Il6-expressing cluster was enriched for Fcer1a (Fc epsilon RI), Cd200r3 (CD200R3), and Mcpt8 (Mast cell protease 8) expression, identifying the cells as a mast/basophil subset (Fig. 4J and fig. S5C). Analysis of several other detectable cytokines indicated that the Il6 expression pattern was distinct in our dataset. For instance, Il1b and Il18 expression was mainly detected in neutrophils and macrophages, respectively (Fig. 4J). Consistent with previous studies showing that human and mouse HSC/progenitors respond to IL-6 (61, 62), expression of genes encoding the IL-6R complex, Il6ra and gp130 (Il6st), were detected in the “progenitor” cluster, and their expression was upregulated in response to C. albicans colonization (fig. S5, D and E). These results indicate that granulopoiesis induced by C. albicans colonization is associated with the expansion of IL-6+ myeloid cells, particularly mast/basophils, in the BM.
Hypha-associated candidalysin promotes granulopoiesis
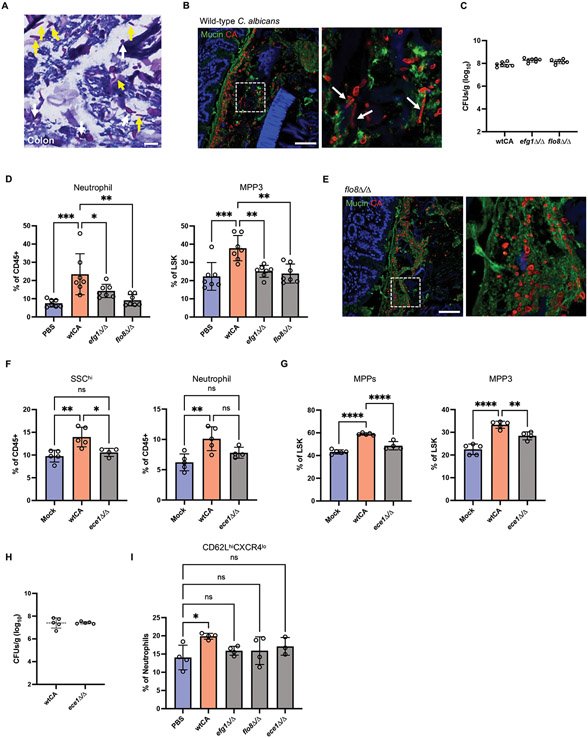
C. albicans morphogenesis and associated virulence factors play important roles in the host immune response and commensal fitness (63-65). Histological analyses of C. albicans in mouse intestines with Periodic acid-Schiff (PAS) staining, which stains polysaccharides on the surface of C. albicans, showed plenty of oval-shaped cells that indicated the typical yeast morphology (Fig. 5A). Elongated morphological structures were also visible. Similar results were obtained by immunofluorescence microscopy (Fig. 5B). A large quantity of yeast-like C. albicans were detected in the mucus layer close to the intestinal epithelium, but excluded from the tissue, suggesting lack of invasion.
Fig. 5. Candidalysin promotes granulopoiesis.
(A) PAS-stained colonic section from C. albicans-colonized antibiotic-treated mice. White and yellow arrows indicate yeast and hyphal structures, respectively. Scale bar represents 10 μm. (B) Confocal images of wild-type C. albicans colonized colon sections immunostained with anti-candida antibody and counterstained with FITC-lectins mixture. Region of interest is magnified in right panel. Arrows indicate the hyphal structure. Scale bar represents 100 μm. (C) Fungal CFUs in feces from antibiotic treated mice 21 days post-inoculation with wild-type C. albicans and yeast-locked mutants. (D) Frequency of neutrophils in the peripheral blood and MPP3 in the BM on day 21 after wild-type and mutant (efg1Δ/Δ and flo8Δ/Δ) C. albicans inoculation. N = 7 mice per group. (E) Confocal images of colon sections from mice colonized with the FLO8 deletion mutant immunostained with anti-candida antibody and counterstained with FITC-lectins mixture. Region of interest is magnified in right panel. Scale bar represents 100 μm. (F) Frequency of SSChi granulocytes and neutrophils in the peripheral blood on day 21 after wild-type and mutant (ece1Δ/Δ) C. albicans inoculation. (G) Frequency of MPPs and MPP3 in the BM on day 21 after wild-type and mutant (ece1Δ/Δ) C. albicans inoculation. N = 5 PBS control, N = 5 wtCA-colonized mice, and N = 4 mutant (ece1Δ/Δ) CA colonized mice. (H) Fungal CFUs in feces from antibiotic treated mice 21 days post-inoculation with wild-type and ece1Δ/Δ C. albicans. (I) Frequency of fresh neutrophils (CD62LhiCXCR4lo) in the peripheral blood on day 21 after wild-type and mutant C. albicans inoculation. Dots in bar graphs correspond to individual mice. Mean and SD are shown. *p < 0.05, **p < 0.01, ****p < 0.0001 by ordinary one-way analysis of variance (ANOVA) with Holm-Sidak multiple comparisons test. ns, not significant.
The transcription factors EFG1 and FLO8 are essential for filamentous growth and regulate the expression of virulence factors in response to environmental cues, such as temperature, pH, or nutrients (66-69). Thus, to examine whether these morphogenic regulators of C. albicans were required for promoting granulopoiesis in the BM, we compared mice colonized with wild-type versus efg1Δ/Δ and flo8Δ/Δ mutant C. albicans. Although these yeast-locked mutants displayed similar burden in stool as the isogenic wild-type control, both efg1Δ/Δ and flo8Δ/Δ C. albicans lost the ability to induce MPP3 in the BM and neutrophils in the blood (Fig. 5, C and D). We confirmed that structures indicative of hyphal morphology were absent in mice colonized by flo8Δ/Δ C. albicans (Fig. 5E). Among the most characterized factors that are upregulated during hyphal formation in C. albicans is the immunogenic mycotoxin candidalysin (70). The candidalysin-deficient (ece1Δ/Δ) mutant did not increase granulocyte and neutrophil levels compared with mock-treated mice. Furthermore, ece1Δ/Δ C. albicans induced significantly reduced MPPs and MPP3 compared with wild-type C. albicans. Fungal burden in stool of wild-type and ece1Δ/Δ C. albicans were comparable (Fig. 5, F-H). In addition, the proportion of newly released neutrophils (CD62LhiCXCR4lo) in mice colonized by these C. albicans mutants were similar to control uninoculated mice (Fig. 5I). These findings indicate that the hypha-associated virulence factor ECE1 promotes granulopoiesis.
Fungal colonization enhances protection against gram-positive bacterial infections
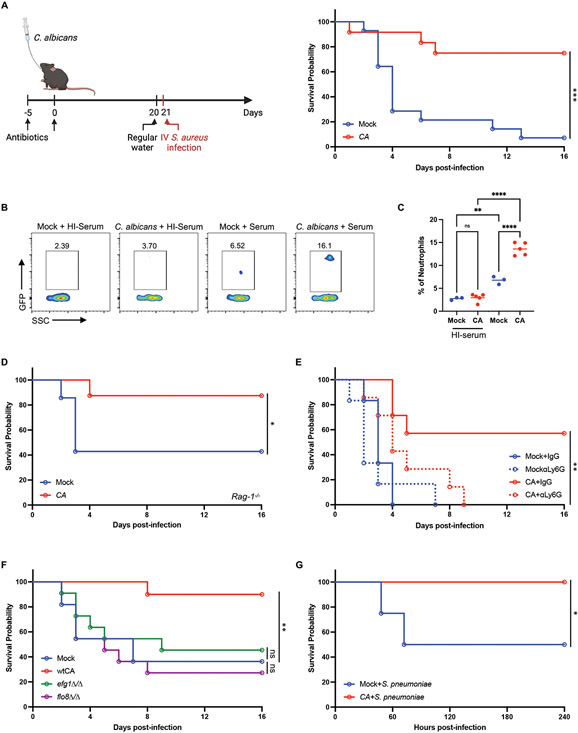
We next asked whether the response to fungal colonization of the gut leads to altered susceptibility to bloodstream infection by S. aureus, a bacterial pathogen sensitive to granulocytes (71). Consistent with this possibility, laboratory mice colonized with C. albicans for 3 weeks displayed substantially enhanced survival following intravenous infection by methicillin resistant S. aureus (MRSA) strain USA300 (Fig. 6A). Our experiments examining cell surface markers suggest neutrophils from C. albicans-colonized mice were qualitatively different. To examine phagocytic capacity ex vivo, we co-cultured neutrophils and GFP-labeled S. aureus incubated with serum to allow for opsonization, and used heat-treated serum in which complement proteins were inactivated as a control. Neutrophils isolated from C. albicans-colonized mice exhibited enhanced phagocytosis of opsonized S. aureus (Fig. 6, B and C), indicating that fungal colonization boosted both quantity and function of neutrophils. Additionally, we found S. aureus burden in various organs were decreased in C. albicans-colonized mice compared with control mice, especially in the blood and lungs (fig. S6A). Fungal colonization has been shown to contribute to expansion of protective Th17 cells and induce an anti-fungal IgG response against pathogens (30, 72). To rule out potential cross-reactivity of lymphocytes between microbes, RAG-1 deficient mice, lacking mature B and T lymphocytes, were challenged with S. aureus after 3 weeks of C. albicans colonization. C. albicans colonized RAG-1 deficient mice, which we confirmed showed comparable fungal burden in the stool as other experiments, displayed improved survival during S. aureus infection compared with the non-colonized control Rag-1−/− mice (Fig. 6D and fig. S6B). In contrast, depletion of neutrophils with anti-Ly6G antibodies abrogated the protection conferred by C. albicans colonization (Fig. 6E). Although the degree of survival among control groups was variable, which may be inherent to sepsis models (73), these results show that C. albicans consistently protected the host across experiments when neutrophils are present. Also, C. albicans mutants that are unable to induce MPP3 in the BM and neutrophils in the blood, efg1Δ/Δ and flo8Δ/Δ, completely lost the ability to protect the mice from S. aureus infection (Fig. 6F and fig. S6C). Lastly, the protection induced by fungal colonization was not specific for S. aureus infection; C. albicans colonized mice also improved survival following intraperitoneal or intranasal infection with Streptococcus pneumoniae (Fig. 6G and S6D). Together, these results show that immune alterations associated with C. albicans colonization of the gut enhance protection against Gram-positive bacterial pathogens.
Fig. 6. Intestinal colonization by C. albicans protects against gram-positive bacterial infections.
(A) Survival following i.v. injection of S. aureus on day 21 following PBS (mock, N=14) or C. albicans (N=12) inoculation. Antibiotics-containing water was swapped with regular water 24 hours before S. aureus infection. 3 independent repeats. (B) Representative flow cytometry plots of neutrophils isolated from the BM of mock and C. albicans-colonized mice incubated with GFP-labeled S. aureus together with untreated or heat-inactivated (HI) mouse serum for 20 minutes at a multiplicity of infection (MOI) of 25. (C) Quantification of frequency of GFP+ neutrophils from (B). N = 3 mock and N = 5 C. albicans-colonized mice. (D) Survival of Rag1−/− knockout mice infected with S. aureus on day 21 after PBS (N=7) or C. albicans (N=8) inoculation. 2 independent repeats. (E) Survival of mice infected with S. aureus on day 21 after PBS or C. albicans inoculation and treated with anti-Ly6G or IgG isotype control antibodies on day −1, 1, 3, 5, and 7 days post-infection. N = 7 mice per group. (F) Survival of mice infected with S. aureus on day 21 after inoculation with PBS, wild-type, or mutant C. albicans (efg1Δ/Δ and flo8D/flo8Δ/Δ). N = 22 mice per group. 2 independent repeats. (G) Survival of mice injected i.p. with S. pneumoniae on day 21 after PBS or C. albicans inoculation. N = 12 mice per group. Dots in bar graphs correspond to individual mice. Mean and SD are shown. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by ordinary one-way analysis of variance (ANOVA) with Holm-Sidak multiple comparisons test (C). (A, D-G) log-rank Mantel–Cox test. ns, not significant.
Intestinal fungal colonization mediates longer-lasting protection compared with innate immune training
Our finding that C. albicans improves survival of Rag1−/− mice in a manner dependent on neutrophils during S. aureus infection indicates that fungal colonization mediates protection by enhancing innate immunity. β-glucan, a potent stimulant of trained immunity, reprograms the immune cells epigenetically or functionally to enhance host responses (74). Trained immunity stimulated by β-glucan induces impressive broad-spectrum protective responses against secondary infections (38, 39, 75). Although our results showed that granulopoiesis wanes over time in transiently stimulated mice compared with persistent fungal colonization (Fig. 3), BM cells from mice injected with β-glucan display a remarkably enhanced ability to reinitiate myeloid cell differentiation upon rechallenge, indicating that some immune changes are sustained in this model (76). Because persistent exposure to fungi maintains granulocyte levels in the blood (Fig. 3), we investigated whether fungal colonization would afford greater protection from a bacterial infection compared with the previously described β-glucan-induced trained immunity that involves a recall response. To test our hypothesis, we compared laboratory mice colonized by C. albicans through oral inoculation and mice injected intraperitoneally with β-glucan that received a secondary challenge with S. aureus either at 7 days or 3 weeks post-colonization or –injection (Fig. 7A). β-glucan-induced circulating neutrophil levels return to baseline at these time points (Fig. 3D); therefore, any protection can be attributed to innate immune memory. At the 7 day time point, C. albicans colonization and β-glucan injection led to a similar degree of enhanced protection compared with control mice (Fig. 7B). In contrast, at 3 weeks post-treatment, protection against S. aureus induced by β-glucan was absent, yet fungal colonization was capable of sustaining enhanced immunity (Fig. 7, C and D). Consistent with this finding, intestinal C. albicans protected Dectin-1−/− mice from S. aureus infection (Fig. 7E). Overall, fungal gut colonization led to sustained changes to the host immune system that mediated greater and prolonged protection against pathogenic infection.
Fig. 7. Intestinal C. albicans colonization mediates longer-lasting protection from S. aureus compared with β-glucan stimulation.
(A) Schematic depicting experimental procedure in (B). Antibiotic-treated mice were orally inoculated with PBS (mock) (N=7) or C. albicans (N=9) or intraperitoneally injected with β-glucan (1mg) (N=9). Antibiotics-containing water was swapped with regular water on day 6 and mice were injected i.v. with S. aureus on day 7. (B) Survival of mice infected with S. aureus on day 7 post-inoculation with C. albicans or β-glucan. (C) Schematic depicting experimental procedure in (D) and (E). Similar to (A) except mice were injected i.v. with S. aureus on day 21. (D) Survival of mice infected with S. aureus on day 21 post-inoculation with C. albicans or β-glucan. N = 25 mice per group. 2 independent repeats. (E) Survival of Dectin-1−/− mice infected with S. aureus on day 21 post-inoculation with C. albicans or β-glucan. N = 7 mice per group. *p < 0.05, **p < 0.01 by log-rank Mantel–Cox test (B, D-E). ns, not significant.
DISCUSSION
Granulocyte abundance and function is one of the most recognized differences between humans and mice. Granulocytes make up only 5-10% of leukocytes in mouse blood but 50-70% in humans. Our results indicate the environment, specifically sustained stimulation by intestinal fungi, may account for at least part of this difference by regulating granulopoiesis in the BM. A comprehensive study showed that gut fungal communities in SPF C57BL/6J laboratory mice vary across vendors and are shaped by the environment, bacterial communities, and diet (77). Although the degree of fungal colonization and community composition is likely facility-dependent, most studies do not report the retrieval of fungi from SPF mice through culturing techniques. Therefore, fungal burden may be low in many conventional animal facilities, as we previously observed (16). In addition to our report, another group showed that wild mice have significantly higher relative abundance of intestinal fungi (13). Thus, a certain level of fungal colonization may be the norm for free-living mammals. Consistent with this idea, the effect of rewilding on granulocytes and acquisition of intestinal fungi was reproducible across years. However, in contrast to the carefully controlled laboratory experiments in which mice are inoculated with a defined amount of C. albicans, we consistently observed greater variance in granulocyte numbers among rewilded mice. We previously showed a positive correlation between granulocyte and fungal abundance, and the specific association between neutrophils and fungal burden was confirmed here. Considering that humans display interindividual differences in the fungal microbiota, including the presence and abundance of pro-inflammatory C. albicans strains (78, 79), inter-individual variation in fungal exposures and immune traits may be another aspect of human biology that is captured in the outdoor mouse enclosure (80).
We note that fungal colonization of mice falls short of fully recapitulating the relative proportion of various leukocytes observed in humans, and that many environmental and species-intrinsic factors likely contribute to deficiencies in the laboratory mouse model for studying human immunity. Exposure to pathogens, as opposed to commensals, may be particularly important for the maturation of certain aspects of the adaptive immune system (9, 12). We also note that the pronounced eosinophil expansion observed in rewilded mice was not reproduced with C. albicans inoculation. Interestingly, fungi isolated from rewilded mice better recapitulated the increase in peripheral eosinophils, suggesting that even among fungi, the exact host response to intestinal exposure is dependent on the fungal species and strains.
Mechanistic insight came from our finding that transcription factors controlling hyphal morphogenesis were necessary for enhanced granulopoiesis, leading us to identify candidalysin as an essential factor in this host response. Candidalysin induces membrane damage in target cells to evoke cytokines including IL-6 and IL-1β (70). Other fungi produce mycotoxins, which may explain why exposure to fungi in the outdoor enclosure induces similar (but not identical) outcomes as colonization by C. albicans. Additionally, fungal sensors such as Dectin-1 were dispensable, further supporting a model in which granulopoiesis is driven by subtle cellular damage by candidalysin, although our findings do not rule out a role for orthogonal pathways that are triggered by yeast-to-hyphae transition. Multiple pathogen sensors act in an overlapping or redundant manner during innate immune memory and Th17 response elicited by intravenous injection and gut colonization with C. albicans, respectively (81, 82), and although β-glucan has received the bulk of attention, a recent study showed that fungal mannans travel through the lymph from the periphery to stimulate a potent CARD9-independent immune response (83).
The functional consequences of the enhanced granulopoiesis in response to fungal colonization was profound. C. albicans can exacerbate the adverse consequences of S. aureus infection when found together, such as through fungal hypha-associated adhesions factors that promote mixed biofilm formation (84-86). We found that when colonization was restricted to the gut, even after a month following the initial inoculation, C. albicans continued to sustain high levels of peripheral neutrophils and substantially protected against bacterial infection. This observation motivated us to compare gut colonization with transient stimulation of antifungal defense, an established trigger of granulopoiesis. Similar to the short-term response to systemic infection by C. albicans (87), we found that neutrophils expanded and declined rapidly in the periphery in a 3-day span after β-glucan injection. Despite this return to baseline, we reproduced observations in the literature by showing that β-glucan injection provided enhanced protection through trained immunity when the bacterial challenge occurred a week after priming. However, β-glucan injection failed to confer protection at one month post priming. The recall response is potentially slower or less efficient at later time points when innate immune memory wanes, whereas mice experiencing persistent fungal colonization benefit from constitutively high levels of circulating fresh neutrophils. It is also possible that reprogramming leads to a qualitatively different response that relies on monocytes and macrophages more so than neutrophils (38, 39). Additionally, the fungal determinants of short versus long term effects on the host are distinct. When delivered intravenously, C. albicans defective in hypha-associated gene expression was able to confer short-term protection from subsequent infectious disease challenges similar to β-glucan injection (27). In contrast, we found that protection conferred by gut colonization was dependent on the hypha-associated gene expression program.
In conclusion, our results indicate that fungal colonization continually promotes granulopoiesis and neutrophil expansion through a mechanism distinct from infection-induced emergency granulopoiesis or trained immunity. The sustained immune activation offers prolonged protection against bloodstream pathogens. Recent evidence highlights a previously unappreciated heterogeneity of neutrophils, which may be key for their function during infectious and chronic diseases, such as cardiovascular inflammation and cancer (88-90). The newly produced neutrophils induced by fungi may carry distinct characteristics that are missing in laboratory mice. Given that humans and free-living mammals are exposed to fungi from the environment constantly, introducing fungi into laboratory mice is likely to better recapitulate certain aspects of mammalian biology.
MATERIALS AND METHODS
Study design
The aim of this study was to reveal how intestinal fungi modulate hematopoietic stem cells and progenitors, particularly myeloid-biased MPP3, leading to increased granulopoiesis and circulating neutrophils in the blood. In addition to rewilding in which laboratory mice experience a semi-natural environment through release into an outdoor enclosure facility, we used human commensal fungi, C. albicans, in the lab to recapitulate the expansion of granulocytes. MPPs and MPP3 in bone marrow were analyzed by flow cytometry to determine the levels of granulopoiesis. β-glucan injection, which mimics fungal infection, was used to compare to fungal colonization in the gut. Lastly, we assessed their protective potential against Gram-positive bacteria. Mice were randomly assigned to individual groups and analyzed without excluding outliers. The rewilding procedure and facility is described in detail below. All laboratory experiments were performed at least two times independently. The number of mice and statistics used in the studies are included in the figure legends.
Mice and wild enclosure
All mouse lines were on a C57BL/6J background and bred onsite in an MNV/ Helicobacter-free specific pathogen free (SPF) facility at NYU Grossman School of Medicine. C57BL/6J (WT), Dectin-1−/− (Clec7a−/−) and Card9−/− mice were initially purchased from The Jackson Laboratory. Rorc(γt)-EGFP (Rorc−/−) mice were obtained from Dr. Dan Littman, NYU Grossman School of Medicine. All animal work was approved by NYU Langone IACUC (#IA16-0087 and #IA16-00864). For experiments performed within the institutional SPF facility that involved inoculation with microbes, 6-10 week old mice of both sexes were used and compared with control-treated littermates. Littermates of mice used for rewilding experiments were generated from multiple breeding pairs and randomly assigned to either remain in the institutional vivarium (laboratory mice) or released into the outdoor enclosures (rewilded mice) to control for the microbiota at the onset of the experiment. Outdoor enclosures were previously described (15, 16) and the protocols for releasing the laboratory mice into the outdoor enclosure facility were approved by Princeton IACUC (#1982-17).
The Stony Ford outdoor enclosure was previously described (15, 16). Briefly, the enclosure consists of wedges, each measuring about 180 m2 and fenced by 1.5-m high, zinced iron walls that are buried >80 cm deep and topped with electrical fencing to keep out terrestrial predators. A straw-filled shed is provided in each enclosure, along with two watering stations and a feeding station, so that the same mouse chow used in the laboratory (PicoLab Rodent Diet 20) was provided ad libitum to all mice. Mice outdoors, however, also had access to food sources found within the enclosures, including berries, seeds, and insects. 12 WT, 10 Dectin-1−/−, and 8 Card9−/− female laboratory mice were housed in the enclosure for 6-7 weeks. 9 WT, 8 Dectin-1−/−, 8 Card9−/− matched littermates were maintained in the institutional vivarium for comparison. Longworth traps baited with chow were used to catch mice after release. All rewilded mice were caught in the final trapping for terminal analyses and all lab control mice were recovered. Euthanasia was performed by CO2 asphyxiation, and blood, femur, and intestinal tissue were harvested. The presence of macroscopic parasites in multiple organs and microscopic parasites by histology were analyzed to confirm that the mice in the outdoor enclosure were free from parasite infection in rewilding experiments.
Construction of C. albicans strains
YPD medium was prepared as previously described (91). SCM medium was prepared as for SCD medium (91) except 2% maltose was included in place of dextrose. YPD supplemented with 200 μg/mL nourseothricin (Werner BioAgents, Jena, Germany) was used to select for nourseothricin-resistant (NATR) strains.
To generate an efg1Δ/Δ strain, pRB721 (92) was digested with Apa I and Sac I and transformed into SC5314 strain to generate NATR EFG1 heterozygotes. Integration was checked by PCR using oligos 2284/4438 and 2286/4439. The SAT1 marker was recycled by growing on SCM medium and a second round of transformation performed to delete the remaining EFG1 allele using pRB721 to create a NATR efg1 null. Correct integration was confirmed and the absence of the EFG1 gene established by PCR using oligos 819/828. The SAT1 marker was recycled by growth on maltose to generate the NAT-sensitive (NATs) efg1 null strain CAY10195.
To delete the FLO8 gene, a pSFS-FLO8 knockout plasmid (pRB989) was first constructed. Oligos 4988/4989 and 4990/4991 were used to PCR amplify 5’ and 3’ UTRs of the FLO8 gene from the SC5314 strain background. These two fragments were cloned into pSFS2A (93) using Apa I/Xho I and Sac I/Sac II sites, respectively. To create a flo8Δ/Δ strain, pRB989 was cut by Apa I/Sac I and transformed into SC5314, generating a FLO8/flo8Δ strain. PCR was performed to check integration using oligos 4982/4438 and 5076/4439. After recycling the SAT1 marker by growth on SCM, pRB989 plasmid was again digested by Apa I/Sac I to perform the second round of transformation, resulting a flo8/flo8 mutant. PCR checks were performed using oligos 4982/4438, 5076/4439 and 5200/4986. The NATR marker was recycled by growth on SCM to generate the NATS flo8Δ/Δ strain CAY10993.
To generate SC5314 ece1Δ/Δ, oligos 4248/4249 were used to amplify the ARG4 and HIS1 cassettes from a published ece1Δ/Δ and transformed into strain SN95 (94). Cells were grown on SC medium without arginine to select for transformants. Junction PCR checks were performed using oligos 4252/4287 and 4286/4253. The transformation was repeated to delete the second ECE1 allele and transformants grown on SC medium lack of histidine and arginine. Transformants were PCR checked using oligos 4250/4251 for ORF, 4252/4289 for the 5’ junction and 4288/4253 for the 3’ junction. To generate the NATR version of ece1Δ/Δ, the pDis3 plasmid was used to integrate into the NEUT5L locus as described previously (95). The plasmid was linearized by NgoMIV and transformed into ece1Δ/Δ to create NATR CAY11507. PCR checks were performed using oligos 3055/3056.
Candida albicans inoculation
A single colony of wild-type or mutant C. albicans was cultured in 5ml of Sabouraud Dextrose (SD) media with chloramphenicol (25 μg/mL, Sigma) at 30°C for 16 h. Cells were pelleted at 1000g for 5 minutes and washed with 1x sterile PBS three times. C. albicans then was re-suspended in 2ml of 1 x PBS. Germ-free or antibiotics-treated (1g of ampicillin and 0.5g of streptomycin in 1L of sterilized water) mice were orally gavaged with 150 μl of C. albicans (~2 × 107 CFUs per mouse) using plastic feeding tubes (Instech Laboratories). After the inoculation of C. albicans, feces and organs were collected and analyzed at different time points according to the experiments. For depletion of colonized C. albicans, germ-free mice were supplemented with fluconazole (0.5 g/L) in drinking water for 2 weeks. The fecal pellets were homogenized in PBS and serial dilutions of fecal samples were plated on SD plates to determine the fungal CFU of wild-type and mutant strains.
Isolation of mouse lamina propria (LP) cells
LP cells were harvested as previously described (Dallari et al., 2021). Briefly, small intestine and colonic tissues were cut open and washed with PBS, and fat was removed. The tissues were incubated first with 20 ml of Hank’s Balanced Salt Solution (HBSS, Gibco) with 2% HEPES, 1% sodium pyruvate, 5 mM Ethylenediaminetetraacetic acid (EDTA), and 1mM DL-dithiothreitol (Sigma-Aldrich) for 15 min at 37°C with 220 rpm and then with new 20 mL of HBSS with 2% HEPES, 1% sodium pyruvate, 5mM EDTA for 10 min at 37°C with 220 rpm. The tissue bits were washed with HBSS, minced, and then enzymatically digested with collagenase (0.5 mg/ml; Sigma-Aldrich) and Deoxyribonuclease I (0.01 mg/ml, Sigma-Aldrich) for 30 min at 37°C with 200 rpm. Digested solutions were passed through 70 μm nylon mesh (ELKO Filtering) and isolated cells were resuspended in 40% Percoll (Sigma-Aldrich), layered onto 80% Percoll, and centrifuged without brake at room temperature at 2,200 rpm for 22 min. Cells were recovered from the interphase and washed with RPMI1640 (Corning) containing 10% fetal bovine serum (FBS, Peak Serum) and used as lamina propria cells.
Flow cytometry analysis
Whole blood was collected through cardiac puncture from mice and kept in a heparin containing tube on ice. Femur was isolated from the surrounding tissue and cleaned with a surgical scalpel and paper towels to remove surrounding muscle and connective tissue. The bone marrow (BM) content was flushed out and BM fluid was collected and kept at −80°C until all samples were collected and analyzed together. 200 μl of blood sample and all BM cells were re-suspended in 5 ml and 2.5 ml of 1x RBC lysis buffer (BioLegend, CA) respectively for 10 minutes to lyse the red blood cells. After two washes with cold PBS, BM and blood cells were signalized through 35 μm strainer with ice cold FACS buffer (PBS, 2% FBS, 1mM EDTA). Single-cell suspensions of BM cells in FACS buffer was counted using an automatic cell counter with trypan blue (Countess 3, ThemoFisher). The staining antibodies for flow cytometry were purchased from Thermo Fisher Scientific, BioLegend or BD Biosciences. For cell surface staining, cells were incubated with antibodies at 4 °C for 20 minutes and fixed for 20 minutes at room temperature. All antibodies were used at a dilution of 1:100 unless otherwise noted. For cell surface phenotype analysis, a mouse lineage antibody cocktail (BD Biosciences, 1:10), anti-Siglec-F (E50-2440), anti-B220 (RA3-6B2), anti-NK1.1 (PK136), anti-TCRβ (H57-597), anti-CD3ε (145-2C11), anti-CXCR4 (L276F12), anti-Ly6G (1A8), anti-CD62L (MEL-14), anti-CD45 (30-F11), anti-CD4 (GK1.5), anti-CD8a (53-6.7), anti-CD127 (A7R34), anti-Ly6C (AL-21), anti-CD11b (M1/70), anti-Flt3 (CD135) (A2F10, 1:50), anti-Sca-1 (Ly6A/E) (E13-16L7), anti-CD150 (TC15-12F12.2, 1:50), anti-CD48 (HM48-1), anti-c-Kit (CD117) (2B8), anti-CD115 (AFS98), anti-GR1 (RB6-8C5), and anti-CXCR2 (5E8/CXCR2) were used. For cytokine staining, cell plated in RPMI1640 and stimulated with 1X Cell Stimulation Cocktail (plus transport inhibitors) (eBiosciecne) for 4 h at 37°C. For intracellular staining of cytokines, cells were permeabilized with Intracellular Staining Permeabilization Wash Buffer (Biolegend) at RT for 20 min in the presence of antibodies. For intracellular staining of the transcription factor, cells were permeabilized with the Foxp3/Transcription Factor Staining Buffer Set (BD Biosciences) at RT for 45 min in the presence of antibodies. The following antibodies for intracellular staining were used: anti-IL-6 (MP5-20F3), anti-IL-17A (TC11-18H10.1), anti-RORγt (562894), and anti-GATA3 (L50-823). Dead cells were excluded using Invitrogen Fixable Blue Dead Cell Stain Kit (Thermo Fisher Scientific). Flow cytometry data was acquired on the ZE5 cell analyzer (BIO-RAD) and analyzed on Flowjo v10.8.1.
Cytokine measurements
To collect BM extracellular fluid, BM content in a femoral bone was harvested by centrifugation at 13,000 rpm for 15 seconds and re-suspended with ice cold PBS (50 μl). The supernatant then was collected after pelleting cells by centrifugation at 350g for 5min at 4°C. Cytokines in supernatants were measured using the Mouse IL-6 and IL-1 beta/IL-1 F2 DuoSet ELISA (R&D systems) according to the manufacturer’s instructions.
Cell depletions and cytokine neutralization
To deplete immune cells or neutralize cytokines, antibiotics-treated mice were injected intraperitoneally one day before and every other day after C. albicans inoculation until 7 days post-inoculation with 200 μg of InVivoMab anti-IL-6R (clone 15A7; BioXCell), 100 μg of InVivoMab anti-Ly6G (clone 1A8; BioXCell), InVivoMab anti-CD4 (clone GK1.5, BioXCell), InVivoMab IL-1α (clone ALF-161; BioXCell), InVivoMab IL-1β (clone B122; BioXCell), and InVivoMab GM-CSF (clone MP1-22E9; BioXCell) in 100 μl PBS respectively. Mock or C. albicans control mice received equal volumes of PBS or equal amounts of InVivoMab Rat IgG isotope control antibodies (clone LTF-2; BioXCell). At day 7, mice were euthanized and organs were harvested for further analysis.
Isolation of primary mouse neutrophils and serum
Adult mice were euthanized and whole blood from cardiac puncture was then collected in heparin containing tubes. Designated serum from supernatant was collected after centrifuging at 2000 rpm for 5 minutes and stored at 4°C until future use. The hind limbs of each mouse were washed with an excess of 70% EtOH. Skin was separated until the femurs were exposed, allowing for removal at the proximal hip and distal knee joint. Excess tissue and muscle were removed with paper towels, and clean bones were placed in fresh 1X PBS on ice. The BM cells were flushed out, washed with FACS buffer, and filtered through a 70μm cell strainer. Single-cell suspensions of whole blood cells and BM cells in FACS buffer was counted using an automatic cell counter with trypan blue and placed on ice. Neutrophil enrichment was performed using EasySep Mouse Neutrophil Enrichment Kit (Stem Cell Technology) following manufacturer’s instructions.
Neutrophil phagocytosis assay
GFP-S. aureus (USA300) strain was streaked to single colonies on a TSA plate. Single colonies were inoculated in TSB broth for overnight culture and then subsequently subcultured 1:100 for 3 h in TSB at 37 °C with shaking. Subcultures were centrifugated at 4000rpm and resuspended to final infection concentration (108 cells/ml) in cold PBS. Bacteria was opsonized by mixing 1:1 with fresh mouse serum or heat inactivated control at 37 °C on rotary board for 45 mins. Neutrophils from whole blood and bone marrow were isolated as described above. Neutrophils were seeded at 105 cells/well in RPMI+10% FBS containing 96-well plates. Plates were allowed to rest for 10 minutes on rotary board at 37 °C, opsonized bacteria were then added at a MOI of 20 and the place was returned to the incubator. After 20 mins, the plate was immediately placed on ice for 3 minutes and extensively washed twice with cold PBS. Neutrophils were stained for live/dead with blue reactive dye and neutrophil cell surface markers as previously described.
Imaging of C. albicans in the murine intestine
At 21 days post-inoculation of wild-type C. albicans or mutant strains, mice were euthanized, and segments with feces of the ileum and colon were fixed in the Methanol-Carnoy’s solution (60% (v/v) dry methanol, 30% (v/v) chloroform, 10% (v/v) glacial acetic acid) for 48 h at room temperature. Fixed tissues were processed twice in 100% ethanol for 20 min and twice in xylene for 15 min, and embedded in paraffin wax. The sections of the tissues were first dewaxed with an initial incubation at 60°C for 10 min and two additional incubations in xylene substitute solution. The samples were re-hydrated in the solutions with a descending ethanol gradient (100, 95, 70, and 50%) for 3 min (two times each) and followed by two washes in water. The sections were then ready for PAS (Periodic Acid Schiff) or immunofluorescence staining. For imaging of fungi, after one wash of PBST (0.1% TritonX-100/PBS) for 5 min, the sections were blocked with 5% normal goat serum for 30 min and sequentially incubated with anti-C. albicans (Thermo Fisher) antibody and fluorophore-tagged secondary antibodies. Samples were counterstained with FITC-conjugated UEA-1 (for mucin) for 45 min at 4°C. Finally, sections were mounted with ProLong Diamond (Thermo Fisher) and imaged with Zeiss LSM710.
Single-cell RNA sequencing
Femur bone marrow cells were harvested from 4 PBS-treated and 4 C. albicans-colonized female mice. Cells from individual mouse were hashtagged using the 3′ CellPlex Kit Set A (10x Genomics PN-1000261) according to manufacturer’s protocol and counted using a Bio-Rad TC20 automated cell counter. Single cells were then encapsulated into emulsion droplets using Chromium Controller (10x Genomics). scRNA-seq and cell multiplexing libraries were constructed using Chromium Single Cell 3′ v3.1 Reagent Kit and 3’ Feature Barcode Kit (PN-1000268 & 1000262 respectively) according to the manufacturer’s protocol. Amplified cDNA was evaluated on a Agilent BioAnalyzer 2100 using a High Sensitivity DNA Kit (Agilent Technologies) and final libraries on a Agilent TapeStation 4200 using High Sensitivity D1000 ScreenTape (Agilent Technologies). Individual libraries were diluted to 2nM and pooled for sequencing. Pools were sequenced with 100 cycle run kits (28bp Read1, 8bp Index1 and 91bp Read2) on the NovaSeq 6000 Sequencing System (Illumina).
Single-cell RNA sequencing data processing
The cellranger’s pipeline version 7.0.0 was used to demultiplex cellular barcodes and aligned reads against the mouse genome (mm10 ensemble). Downstream RNA-seq analysis was done with Seurat version 4.0.0 on R version 4.2.1 with the filtered RNA and HTO featured counts. Cells containing more than 20% mitochondrial DNA and less than 300 feature genes were filtered out for initial quality control. HTOs were normalized using centered log-ratio transformation and demultiplexed with the function HTODemux to identify doublets; additional doublets were removed using the scDblFinder package with a calculated doublet rate of 0.14. Regularized negative binomial regression was performed for RNA normalization using the SCTranformation function. Principle component analysis was performed, and the Louvain algorithm determined unsupervised clustering with 20 dimensions. UMAP representation based on totalVI dimension reduction of RNA was used to visualize data. Cell types were determined by a combination of unbiases clustering, canonical cell type marker signatures, and cell type annotation via the SingleR package with the ImmGenData and MouseRNAseqData open-source (expression) reference databases in the CellDex package. The Wilcox test assessed differentially expressed genes between clusters and treatment groups with a Benjamin-Hochberg p-value adjustment.
Bacteria strains and growth conditions
S. aureus strain USA300 LAC clone AH1263 (96), S. pneumoniae P2406 (serotype 4, strain TIGR4; clinical isolate with streptomycin resistance) (97), and S. pneumoniae P2090 (serotype 3; ATCC 6303) were used for in vivo infection studies. S. aureus strains were grown on tryptic soy agar (TSA) or tryptic soy broth (TSB). S. aureus cultures were grown in 5 ml of medium in 15 ml tubes shaking with a 45° angle at 37 °C. For all experiments, S. aureus was grown in TSB overnight and a 1:100 dilution of overnight cultures was sub-cultured into fresh TSB. S. aureus grown to early stationary phase (3 h) was collected and normalized by OD600 for further experimental analysis. S. pneumoniae P2406 and P2090 were grown statically in tryptic soy broth at 37oC to mid-log phase (optical density of 1.0 at 620nm), washed once in Dulbecco’s phosphate buffered saline (dPBS), and diluted to obtain a dose of 100 CFU/gram of body weight in 100μL of dPBS and a dose of 104 CFU in 80μL of dPBS respectively.
In vivo infections
Mice under antibiotics-treatment were inoculated with wild-type C. albicans or mutant strains and colonized for 21 days. 24 hours before bacterial challenge, antibiotics in drinking water was replaced to regular water. For S. aureus infection, mice were anesthetized with Avertin (2,2,2-tribromoethanol dissolved in tert-Amyl alcohol and diluted to a final concentration of 2.5 % vol/vol in 0.9 % sterile saline) by intraperitoneal injection. Mice then were challenged with 1x107 CFUs by retro-orbital injection. Signs of morbidity (>30% weight loss, ruffled fur, hunched posture, paralysis, inability to walk, or inability to consume food or water) were monitored after infection. For S. pneumoniae P2406 infection, mice were inoculated via the intraperitoneal route and monitored daily following infection. For intranasal inoculation, mice were administered S. pneumoniae P2090 following anesthesia with isoflurane and monitored daily.
Quantification and statistical analysis
Statistical parameters including the definition of central value and the exact number (N) of mice per group are annotated in the corresponding figure legend. Results in graphs and bar plots are displayed as mean ± SD. Statistical analysis was performed with GraphPad Prism version 9.2 for Mac (GraphPad) by using an unpaired two-tailed Student’s t test to evaluate differences between two groups. For statistical analysis of multiple groups, the ordinary one-way ANOVA with Holm-Sidak multiple comparisons test was applied. For statistical analysis of survival among groups, log-rank Mantel–Cox test was applied. (* p value < 0.05; ** p value < 0.01; *** p value < 0.001; **** p value < 0.0001; ns, not statistically significant).
Supplementary Material
Acknowledgments:
We wish to thank the NYU Grossman School of Medicine Flow Cytometry and Cell Sorting, Microscopy, Genomic Technology, and Histology Cores (supported by National Institutes of Health [NIH] grants P31CA016087, S10OD01058, and S10OD018338) and Margie Alva, Juan Carrasquillo, and David Basnight of the Gnotobiotics facility for their technical support and use of instruments.
Funding:
This research was supported by National Institutes of Health grants DK093668 (K.C.), AI121244 (K.C. and V.J.T.), HL123340 (K.C.), AI130945 (K.C.), AI140754 (K.C.), DK124336 (K.C.), and DK122698 (F.Y.), AI140754 (K.C. and V.J.T.), and Intramural Research Program of the NIAID, NIH (P.L.). Additional support was provided by the Faculty Scholar grant from the Howard Hughes Medical Institute (K.C.), Crohn’s & Colitis Foundation (K.C., J.A.), Kenneth Rainin Foundation (K.C.), Judith & Stewart Colton Center of Autoimmunity (K.C., J.A.), research station and research rebate awards from PU EEB (A.L.G.), National Science Foundation (A.L.G.). K.C. and V.J.T. are a Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases. This work was also supported by the Bernard Levine Postdoctoral Research Fellowship in Immunology (Y.H.C.) and the Charles H. Revson Senior Fellowships in Biomedical Science (Y.H.C.) and a Cystic Fibrosis Foundation postdoctoral fellowship award LACEY19FO (K.A.L.).
Footnotes
Competing interests: K.C. has received research support from Pfizer, Takeda, Pacific Biosciences, Genentech, and Abbvie. K.C. has consulted for or received an honoraria from Puretech Health, Genentech, and Abbvie. K.C. is an inventor on U.S. patent 10,722,600 and provisional patent 62/935,035 and 63/157,225. V.J.T. has consulted for Janssen Research & Development, LLC, and have received honoraria from Genentech and Medimmune. He is also an inventor on patents and patent applications filed by New York University, which are currently under commercial license to Janssen Biotech Inc. Janssen Biotech Inc. provides research funding and other payments associated with a licensing agreement.
Data and materials availability: The scRNA sequencing data for this study have been deposited in GEO (GSE231824). Requests for fungal strains should be addressed to K.C. (for wild fungi) or R.J.B. (for C. albicans) and should be covered by a material transfer agreement. Requests for Rorc−/− mice should be addressed to the donating investigator. All other genetically engineered mice used in this study are commercially available. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References
- 1.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R, Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Smith K, McCoy KD, Macpherson AJ, Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol 19, 59–69 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Ivanov II et al. , Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill DA et al. , Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol 3, 148–158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lima-Junior DS et al. , Endogenous retroviruses promote homeostatic and inflammatory responses to the microbiota. Cell 184, 3794–3811 e3719 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mestas J, Hughes CC, Of mice and not men: differences between mouse and human immunology. J Immunol 172, 2731–2738 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Tao L, Reese TA, Making Mouse Models That Reflect Human Immune Responses. Trends Immunol 38, 181–193 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Masopust D, Sivula CP, Jameson SC, Of Mice, Dirty Mice, and Men: Using Mice To Understand Human Immunology. J Immunol 199, 383–388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beura LK et al. , Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reese TA et al. , Sequential Infection with Common Pathogens Promotes Human-like Immune Gene Expression and Altered Vaccine Response. Cell Host Microbe 19, 713–719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi YJ et al. , SERPINB1-mediated checkpoint of inflammatory caspase activation. Nat Immunol 20, 276–287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiege JK et al. , Mice with diverse microbial exposure histories as a model for preclinical vaccine testing. Cell Host Microbe 29, 1815–1827 e1816 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosshart SP et al. , Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosshart SP et al. , Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 171, 1015–1028 e1013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung JM et al. , Rapid environmental effects on gut nematode susceptibility in rewilded mice. PLoS Biol 16, e2004108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeung F et al. , Altered Immunity of Laboratory Mice in the Natural Environment Is Associated with Fungal Colonization. Cell Host Microbe 27, 809–822 e806 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin JD et al. , Rewilding Nod2 and Atg16l1 Mutant Mice Uncovers Genetic and Environmental Contributions to Microbial Responses and Immune Cell Composition. Cell Host Microbe 27, 830–840 e834 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Odds FC et al. , Candida albicans strain maintenance, replacement, and microvariation demonstrated by multilocus sequence typing. J Clin Microbiol 44, 3647–3658 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, Sudbery P, Candida albicans, a major human fungal pathogen. J Microbiol 49, 171–177 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Rao C et al. , Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature 591, 633–638 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer FL, Wilson D, Hube B, Candida albicans pathogenicity mechanisms. Virulence 4, 119–128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson DS, Carlisle PL, Kadosh D, Coevolution of morphology and virulence in Candida species. Eukaryot Cell 10, 1173–1182 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witchley JN et al. , Candida albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection. Cell Host Microbe 25, 432–443 e436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doron I et al. , Mycobiota-induced IgA antibodies regulate fungal commensalism in the gut and are dysregulated in Crohn's disease. Nat Microbiol 6, 1493–1504 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang SH et al. , Hemizygosity Enables a Mutational Transition Governing Fungal Virulence and Commensalism. Cell Host Microbe 25, 418–431 e416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ost KS et al. , Adaptive immunity induces mutualism between commensal eukaryotes. Nature 596, 114–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tso GHW et al. , Experimental evolution of a fungal pathogen into a gut symbiont. Science 362, 589–595 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Bacher P et al. , Human Anti-fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida albicans. Cell 176, 1340–1355 e1315 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Jiang TT et al. , Commensal Fungi Recapitulate the Protective Benefits of Intestinal Bacteria. Cell Host Microbe 22, 809–816 e804 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao TY et al. , Commensal Candida albicans Positively Calibrates Systemic Th17 Immunological Responses. Cell Host Microbe 25, 404–417 e406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leonardi I et al. , Mucosal fungi promote gut barrier function and social behavior via Type 17 immunity. Cell 185, 831–846 e814 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atarashi K et al. , Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conti HR et al. , Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med 206, 299–311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirai H et al. , C/EBPbeta is required for 'emergency' granulopoiesis. Nat Immunol 7, 732–739 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Mitroulis I, Kalafati L, Hajishengallis G, Chavakis T, Myelopoiesis in the Context of Innate Immunity. J Innate Immun 10, 365–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manz MG, Boettcher S, Emergency granulopoiesis. Nat Rev Immunol 14, 302–314 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Bekkering S et al. , Metabolic Induction of Trained Immunity through the Mevalonate Pathway. Cell 172, 135–146 e139 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Dos Santos JC et al. , beta-Glucan-Induced Trained Immunity Protects against Leishmania braziliensis Infection: a Crucial Role for IL-32. Cell Rep 28, 2659–2672 e2656 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Moorlag S et al. , beta-Glucan Induces Protective Trained Immunity against Mycobacterium tuberculosis Infection: A Key Role for IL-1. Cell Rep 31, 107634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Netea MG et al. , Trained immunity: A program of innate immune memory in health and disease. Science 352, aaf1098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quintin J et al. , Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 12, 223–232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng SC et al. , mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor PR et al. , The beta-glucan receptor, dectin-1, is predominantly expressed on the surface of cells of the monocyte/macrophage and neutrophil lineages. J Immunol 169, 3876–3882 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Walachowski S, Tabouret G, Fabre M, Foucras G, Molecular Analysis of a Short-term Model of beta-Glucans-Trained Immunity Highlights the Accessory Contribution of GM-CSF in Priming Mouse Macrophages Response. Front Immunol 8, 1089 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leonardi I et al. , CX3CR1(+) mononuclear phagocytes control immunity to intestinal fungi. Science 359, 232–236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adrover JM et al. , A Neutrophil Timer Coordinates Immune Defense and Vascular Protection. Immunity 50, 390–402 e310 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Casanova-Acebes M et al. , Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell 153, 1025–1035 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pietras EM et al. , Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell 17, 35–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson A et al. , Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Evrard M et al. , Developmental Analysis of Bone Marrow Neutrophils Reveals Populations Specialized in Expansion, Trafficking, and Effector Functions. Immunity 48, 364–379 e368 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Goodridge HS, Wolf AJ, Underhill DM, Beta-glucan recognition by the innate immune system. Immunol Rev 230, 38–50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Theel ES, Doern CD, beta-D-glucan testing is important for diagnosis of invasive fungal infections. J Clin Microbiol 51, 3478–3483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carvalho A et al. , Dectin-1 isoforms contribute to distinct Th1/Th17 cell activation in mucosal candidiasis. Cell Mol Immunol 9, 276–286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flannigan KL et al. , IL-17A-mediated neutrophil recruitment limits expansion of segmented filamentous bacteria. Mucosal Immunol 10, 673–684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jouault T et al. , Candida albicans phospholipomannan is sensed through toll-like receptors. J Infect Dis 188, 165–172 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Fuchs K et al. , The fungal ligand chitin directly binds TLR2 and triggers inflammation dependent on oligomer size. EMBO Rep 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagener J et al. , Fungal chitin dampens inflammation through IL-10 induction mediated by NOD2 and TLR9 activation. PLoS Pathog 10, e1004050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarzenberger P et al. , IL-17 stimulates granulopoiesis in mice: use of an alternate, novel gene therapy-derived method for in vivo evaluation of cytokines. J Immunol 161, 6383–6389 (1998). [PubMed] [Google Scholar]
- 59.Stark MA et al. , Phagocytosis of apoptotic neutrophils regulates granulopoiesis via IL-23 and IL-17. Immunity 22, 285–294 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Metcalf D, Hematopoietic cytokines. Blood 111, 485–491 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berardi AC, Wang A, Levine JD, Lopez P, Scadden DT, Functional isolation and characterization of human hematopoietic stem cells. Science 267, 104–108 (1995). [DOI] [PubMed] [Google Scholar]
- 62.Maeda K et al. , Interleukin-6 aborts lymphopoiesis and elevates production of myeloid cells in systemic lupus erythematosus-prone B6.Sle1.Yaa animals. Blood 113, 4534–4540 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mukaremera L, Lee KK, Mora-Montes HM, Gow NAR, Candida albicans Yeast, Pseudohyphal, and Hyphal Morphogenesis Differentially Affects Immune Recognition. Front Immunol 8, 629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gantner BN, Simmons RM, Underhill DM, Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J 24, 1277–1286 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.d'Enfert C et al. , The impact of the Fungus-Host-Microbiota interplay upon Candida albicans infections: current knowledge and new perspectives. FEMS Microbiol Rev 45, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao F et al. , The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol Biol Cell 17, 295–307 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sohn K, Urban C, Brunner H, Rupp S, EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol Microbiol 47, 89–102 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Lane S, Birse C, Zhou S, Matson R, Liu H, DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J Biol Chem 276, 48988–48996 (2001). [DOI] [PubMed] [Google Scholar]
- 69.Hollomon JM et al. , The Candida albicans Cdk8-dependent phosphoproteome reveals repression of hyphal growth through a Flo8-dependent pathway. PLoS Genet 18, e1009622 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moyes DL et al. , Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532, 64–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rigby KM, DeLeo FR, Neutrophils in innate host defense against Staphylococcus aureus infections. Semin Immunopathol 34, 237–259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doron I et al. , Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell 184, 1017–1031 e1014 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arrazuria R et al. , Variability of murine bacterial pneumonia models used to evaluate antimicrobial agents. Front Microbiol 13, 988728 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fanucchi S, Dominguez-Andres J, Joosten LAB, Netea MG, Mhlanga MM, The Intersection of Epigenetics and Metabolism in Trained Immunity. Immunity 54, 32–43 (2021). [DOI] [PubMed] [Google Scholar]
- 75.Ciarlo E et al. , Trained Immunity Confers Broad-Spectrum Protection Against Bacterial Infections. J Infect Dis 222, 1869–1881 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mitroulis I et al. , Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell 172, 147–161 e112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mims TS et al. , The gut mycobiome of healthy mice is shaped by the environment and correlates with metabolic outcomes in response to diet. Commun Biol 4, 281 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li XV et al. , Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature 603, 672–678 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iliev ID, Cadwell K, Effects of Intestinal Fungi and Viruses on Immune Responses and Inflammatory Bowel Diseases. Gastroenterology 160, 1050–1066 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Graham AL, Naturalizing mouse models for immunology. Nat Immunol 22, 111–117 (2021). [DOI] [PubMed] [Google Scholar]
- 81.Reales-Calderon JA et al. , Gut-Evolved Candida albicans Induces Metabolic Changes in Neutrophils. Front Cell Infect Microbiol 11, 743735 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shao TY et al. , Candida albicans oscillating UME6 expression during intestinal colonization primes systemic Th17 protective immunity. Cell Rep 39, 110837 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Borriello F et al. , An adjuvant strategy enabled by modulation of the physical properties of microbial ligands expands antigen immunogenicity. Cell 185, 614–629 e621 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peters BM et al. , Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology (Reading) 158, 2975–2986 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Dyck K et al. , Adhesion of Staphylococcus aureus to Candida albicans During Co-Infection Promotes Bacterial Dissemination Through the Host Immune Response. Front Cell Infect Microbiol 10, 624839 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Todd OA et al. , Candida albicans Augments Staphylococcus aureus Virulence by Engaging the Staphylococcal agr Quorum Sensing System. mBio 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Basu S et al. , "Emergency" granulopoiesis in G-CSF-deficient mice in response to Candida albicans infection. Blood 95, 3725–3733 (2000). [PubMed] [Google Scholar]
- 88.Silvestre-Roig C, Braster Q, Ortega-Gomez A, Soehnlein O, Neutrophils as regulators of cardiovascular inflammation. Nat Rev Cardiol 17, 327–340 (2020). [DOI] [PubMed] [Google Scholar]
- 89.Eruslanov EB, Singhal S, Albelda SM, Mouse versus Human Neutrophils in Cancer: A Major Knowledge Gap. Trends Cancer 3, 149–160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hedrick CC, Malanchi I, Neutrophils in cancer: heterogeneous and multifaceted. Nat Rev Immunol 22, 173–187 (2022). [DOI] [PubMed] [Google Scholar]
- 91.Guthrie C, Fink G, Guide to Yeast Genetics and Molecular Biology. (Academic Press, 1991). [Google Scholar]
- 92.Liang SH et al. , Hemizygosity enables a mutational transition governing fungal virulence and commensalism. Cell Host Microbe 25, 418–431.e416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reuss O, Vik A, Kolter R, Morschhauser J, The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341, 119–127 (2004). [DOI] [PubMed] [Google Scholar]
- 94.Noble SM, Johnson AD, Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4, 298–309 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gerami-Nejad M, Zacchi LF, McClellan M, Matter K, Berman J, Shuttle vectors for facile gap repair cloning and integration into a neutral locus in Candida albicans. Microbiology (Reading) 159, 565–579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boles BR, Thoendel M, Roth AJ, Horswill AR, Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5, e10146 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zafar MA, Kono M, Wang Y, Zangari T, Weiser JN, Infant Mouse Model for the Study of Shedding and Transmission during Streptococcus pneumoniae Monoinfection. Infect Immun 84, 2714–2722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fonzi WA, Irwin MY, Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717–728 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.